* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Gene expression clustering using gene ontology and biological
Epigenetics in stem-cell differentiation wikipedia , lookup
Point mutation wikipedia , lookup
Transposable element wikipedia , lookup
Oncogenomics wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
X-inactivation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Copy-number variation wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Long non-coding RNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
Public health genomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Genomic imprinting wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Genome evolution wikipedia , lookup
Ridge (biology) wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genome (book) wikipedia , lookup
Gene therapy wikipedia , lookup
The Selfish Gene wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene desert wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Gene nomenclature wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Microevolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Designer baby wikipedia , lookup
Gene expression profiling wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
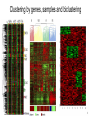
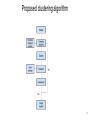
Gene expression clustering using gene ontology and biological networks Student: Peter Leontev, SPbAU Scientific advisor: Son Pham, UCSD Gene expression data source ● Microarrays ● RNA-seq ● Single-cell RNA-seq 2 Gene expression data representation ● CPM, FPKM, RPKM, TPM ● Heatmap/PCA/tSNE/... 3 Clustering in a nutshell ● Similar items should fall into the same clusters whereas dissimilar items should fall in different clusters. ● There is no single best criterion for obtaining a partition because no single and precise definition of cluster exist. ● As a result, there are tremendous amount of methods. 4 Proximity measure ● Gene expresion values can be formalized as numerical vectors: ● There are many proximity metrics such as L1 and L2 norms, Mahalanobis distance, correlation, etc. 5 Clustering by genes, samples and biclustering 6 Two popular clustering algorithms Hierchical clustering (HC) allows us to cluster both genes and samples in one picture and see whole dataset. 7 Two popular clustering algorithms K-means is a partitioning method which requires predefined number (K) of clusters. 8 Disadvantages of proximity-based only algorithms ● Use simple distance metrics for large-scale datasets. ● Consider genes as independent entities. ● Ignore known functions of genes. Ref: When Is “Nearest Neighbor" Meaningful? Beyer et al, 1999 9 Embedding prior knowledge ● Many clustering algorithms were developed that use knowledge databases in the clustering process. ● However, they follow the principle that integrating one external source of knowledge to guide an algorithm is enough. 10 Existing approaches (gene ontology) ● A knowledge-based clustering algorithm driven by Gene Ontology, Cheng et al, 2004. ● Seeing the forest for the trees: using the Gene Ontology to restructure hierarchical clustering, Dotan-Cohen et al, 2009. 11 Existing approaches (biological networks) ● A network-assisted co-clustering algorithms to discover cancer subtypes based on gene expression, Liu, 2014. ● Improving clustering with metabolic pathway data, Diego et al, 2015. 12 The goal To combine different biological networks and gene ontology to integrate them into gene expression (bi)clustering algorithm. 13 Proposed clustering algorithm 14 Combining biological networks ● If possible consider any network as directed and weighted. ● Take into account as much as possible edge’s information. 15 Evaluation Hypothesis: gene that show similar expression patterns have common function and so may have the similar networks features too. Evaluation must answer the following questions: 1. How similar gene expression values? 2. Do we have consistency in the gene ontology terms? 3. Can we infer some pathways?(*) 16 Dataset ● Darmanis S, Sloan SA, Zhang Y, Enge M et al. “A survey of human brain transcriptome diversity at the single cell level” (GSE67835). 17 Relevance ● Do we have enough knowledge to cluster gene expression data to determine cell types? ● Do we have enough knowledge to cluster gene expression data to determine cell subtypes? 18 Thank you! Questions? 19