* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Gene regulation I Biochemistry 302
Metalloprotein wikipedia , lookup
RNA silencing wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Non-coding DNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Expression vector wikipedia , lookup
Peptide synthesis wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Proteolysis wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Messenger RNA wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Genetic code wikipedia , lookup
Point mutation wikipedia , lookup
Gene regulatory network wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gene expression wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Biosynthesis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Epitranscriptome wikipedia , lookup
Amino acid synthesis wikipedia , lookup
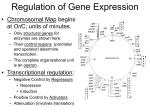
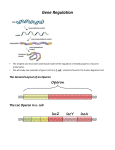
Gene regulation II Biochemistry 302 Bob Kelm February 28, 2005 Catabolic operons: Regulation by multiple signals targeting different TFs Catabolite repression: Activity of lac operon is restricted when both glucose and lactose are present. E. coli would prefer to metabolize glucose directly (via glycolysis) rather than generating it from secondary sugars. CRP homodimer (subunit Mr 22,000) bound to DNA. cAMP “inducer” is in red. RNAP interaction domain is yellow. Lehninger Principles of Biochemistry, 4th ed., Ch 28 Other side of the coin: the biosynthetic trp operon • Amino acid biosynthesis consumes energy – Advantageous to inhibit synthesis of biosynthetic enzymes when end product (amino acid) is available. – Regulatory goal is to repress gene activity. • E. coli trp operon (in contrast to lac) – Trp repressor is activated by ligand (Trp) binding. – Additional regulation by premature termination of transcription (attenuation – regulatory dimmer switch involves ribosome positioning on 5′ mRNA) • Discovered by Charles Yanofsky, common to many biosynthetic operons including Trp, Leu, and His • Dictated by changes in RNA secondary structure • Extends the possible range of transcription rates (moderate to high Trp levels) Schematic of the E. coli trp operon (regulation by Trp-induced repression) chorismic acid → Trp Dimeric HTH protein Trp inducer aporepressor (when trp levels are low) Fig. 26-33 Secondary mechanism of repression: moderate to high Trp levels Structure(s) of the trp operon mRNA leader (trpL) sequence (162 nt) Does the 3:4 pair structure remind you of anything? Lehninger Principles of Biochemistry, 4th ed., Ch 28 Mechanism of transcriptional attenuation Ribosome follows closely behind RNAP as transcription proceeds. The ribosome sterically hinders 2:3 base-pairing upon encountering leader peptide stop codon. Ribosome stalling at Trp codons due to low [Trp-tRNATrp] i.e. when Trp levels are low. This allows more favored 2:3 base-pairing at the expense of 3:4 basepairing. Short leader peptide has no known cellular function. Its synthesis is merely an operon regulatory device. Lehninger Principles of Biochemistry, 4th ed., Ch 28 Another view of attenuation emphasizing importance of the ribosome Fig. 26-36 Regulons: Network of operons with a common regulator • Metabolism of secondary sugars – Lactose, arabinose, and galactose – CRP-cAMP-dependent • Heat-shock gene system – Replacement of σ70 specificity factor by σ32 – RNA polymerase directed to different set of heatshock gene promoters • σ70 σ32 SOS response to DNA damage – LexA repressor – RecA protein (unique role) Lehninger Principles of Biochemistry, 4th ed., Ch 28 Induction of SOS response in E. coli (LexA-dependent regulon) • • • Cellular response to extensive DNA damage Induced genes mostly involved in DNA repair Mechanism: Proteolytic inactivation of LexA repressor – RecA/ssDNA-dependent – Interaction of ssDNA-bound RecA stimulates intrinsic protease activity of LexA. – LecA inactivates itself by catalyzing its own cleavage at a specific Arg-Gly bond in the middle of the protein. Lehninger Principles of Biochemistry, 4th ed., Ch 28 Translation regulation in bacteria: feedback control of ribosomal proteins • Translational feedback in some ribosomal protein (rprotein) operon transcripts – β operon contains genes encoding RNAP subunits – str operon contain genes encoding translational elongation factors • Specific r-proteins possess both rRNA & operon-specific mRNA-binding affinity – Repress translation of operon transcripts when level of r-protein > rRNA – Ensures balanced r-protein and rRNA synthesis Differential binding affinity of L10, S7, S4, L4, and S8 for rRNA (higher) and its owns mRNA transcript (lower) makes this mechanism possible. rRNA synthesis is also regulated by a translation-dependent pathway • • Stringent response: regulation coordinated with [amino acid] Amino acid starvation halts rRNA synthesis by a sequence of events triggered by binding of an uncharged tRNA to ribosome A site then…. – Stringent factor (RelA) binds to ribosome – RelA catalyzes addition of pyrophosphate to 3′ position of GTP then phosphohydrolase removes one phosphate → guanosine tetraphosphate – ppGpp binds to RNA polymerase and alters promoter selectivity (including seven rRNA operons) cAMP and ppGpp are major cellular second messengers in E. coli. Lehninger Principles of Biochemistry, 4th ed., Ch 28