* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Figure 18.19 Regulation of a metabolic pathway
G protein–coupled receptor wikipedia , lookup
Magnesium transporter wikipedia , lookup
Signal transduction wikipedia , lookup
Protein moonlighting wikipedia , lookup
Hedgehog signaling pathway wikipedia , lookup
List of types of proteins wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Gene regulatory network wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
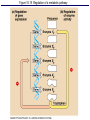
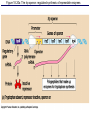
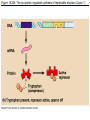
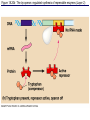
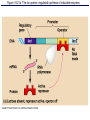
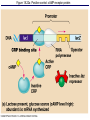
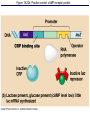
The control of gene expression enable individual bacteria to adjust their metabolism to environmental change If a bacterial cell is deprived of the amino acid, tryptophan which it needs to survive, it will activate its own metabolic pathway to make its own. Figure 18.19 Regulation of a metabolic pathway Operon : consists of a closely related group of genes that act together and code for the enzymes that control a particular metabolic pathway; consists of an operator, promoter, and the genes they control The “switch” for turning the genes off and on is called an operator. It is positioned within the promoter or between the promoter and enzyme-encoding genes, controlling access to the genes. What determines if the operator is on? By itself, it is on – RNA polymerase can bind to the promoter. It can be switched off by a protein called a repressor. The repressor is a product of a regulatory gene. These are transcribed continuously, although at a low rate. Figure 18.20a The trp operon: regulated synthesis of repressible enzymes Figure 18.20b The trp operon: regulated synthesis of repressible enzymes (Layer 1) Figure 18.20b The trp operon: regulated synthesis of repressible enzymes (Layer 2) Tryptophan (trp) is synthesized from E. coli from a precursor molecule in a series of steps. The trp operon is said to be a repressible operon because transcription of it is inhibited when a specific small molecule (tryptophan) binds to a regulatory protein In contrast, an inducible operon, like the lac operon, is stimulated (i.e. induced) when a specific small molecule interacts with a regulatory protein. Figure 18.21a The lac operon: regulated synthesis of inducible enzymes Figure 18.21b The lac operon: regulated synthesis of inducible enzymes The disaccharide, lactose, is available to E. coli if the human host drinks milk. The bacteria can absorb the lactose and break it down for energy. Lactose metabolism begins with the hydrolysis of lactose into its two monosaccharides The enzyme that catalyzes this reaction is called beta galactosidase. In the presence of lactose it can increase 1000x in 15 min. The gene for beta galactosidase is part of an operon, the lac operon, that includes two other genes coding for proteins that function in lactose metabolism Figure 18.22a Positive control: cAMP receptor protein Figure 18.22b Positive control: cAMP receptor protein