* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Gene Regulation in Prokaryotic Cells
Transcription factor wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Minimal genome wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
DNA supercoil wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Non-coding RNA wikipedia , lookup
Molecular cloning wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
DNA vaccination wikipedia , lookup
Genome evolution wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenomics wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Gene expression profiling wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Non-coding DNA wikipedia , lookup
Point mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Primary transcript wikipedia , lookup
Designer baby wikipedia , lookup
Helitron (biology) wikipedia , lookup
Microevolution wikipedia , lookup
Epigenetics of human development wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
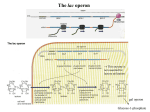
Bio300 Handout #15 Fall, 2004 Gene Regulation in Prokaryotic Cells (Chapter 10) I. The basics of prokaryotic transcriptional regulation A. Key Words: promoter, operator, activator, repressor. The binding of regulatory proteins can either activate or block transcription – Fig. 10-2. B. Domains of DNA binding proteins (activators or repressors) • DNA binding domain • Allosteric site and allosteric effectors allosteric: of, relating to, undergoing, or being a change in the shape and activity of a protein (as an enzyme) that results from combination with another substance at a point other than the chemically active site. (Wester’s Dictionary) allosteric effectors : small molecules interacting with a allosteric site. • Some activator or repressor proteins must bind to their allosteric effectors before they can bind DNA. Others are opposite – loosing the ability to bind DNA once they bind effectors – Fig. 10-3. II. An overview of the lac regulatory circuit in E. coli A. Operon (one promoter controlling several genes) – Fig. 10-4 C. Structural genes (lacZ, lacY and lacA) vs. the regulatory gene (lacI). D. Two DNA docking sites – promoter and operator B. The induction of the lac system. E. The operon is off normally – binding of the lactose-free Lac repressor to the operator sequence. F. Binding of lactose of its analogs to the repressor protein results in an allosteric transition so that the repressor cannot bind the operator DNA anymore. G. A RNA polymerase binds to the promoter and turns on transcription. III. Discovery of the lac system of negative control A. Jacob and Monod found that genes were controlled together. H. Genetic evidence for the presence of gene I, which regulates the expression of gene lacZ encoding β-galactosidase. β-Galactosidase is an inducible enzyme. New enzymes (containing radioactively labeled amino acids) were induced as early as 3 min after the inducer was added. I. Several mutants that couldn’t be complemented by one another – different genes. J. The lacZ, lacY and lacA genes were very closely linked on the chromosome, as shown by recombination mapping. B. Genetic evidence for the operator and repressor K. Used the non-metabolizable induce, isopropyl-β-D-thiogalactoside (IPTG) – Fig. 10-7. L. Assessment of dominance. Created partial diploids by using F’ factors. 1 Bio300 Handout #15 Fall, 2004 M. Cis-acting Oc constitutive mutants – Table 10-1. The wild type lacY gene is cis to the wild type operator and the wild type lacZ gene is cis to the Oc constitutive mutant – Strain 4. N. This cis-acting property of O suggests that it acts simply as a protein-binding site (a piece of DNA) and makes no gene product – Fig. 10-8. O. I+ is dominant to I-; I+ is trans-acting. The I+ gene product can regulate all structural lac operon genes, whether in cis or trans (residing on different DNA molecules) because the protein product of the I gene is able to diffuse and act on both operators in the partial diploid – Table 10-2, Fig. 10-9. C. Genetic evidence for allostery P. Suppressor (Is) mutations. Is repressors cannot bind the IPTG inducer, but are still able to bind to the operator – Table 10-3, Fig. 10-10. Q. Is mutation are dominate to I+. So the repression is constitutive; cannot be derepressed by IPTG. VI. Catabolite repression of the lac operon: positive control A. Choosing the best sugar to metabolize R. A breakdown product, or catabolite, of glucose prevents activation of the lac operon by lactose. S. In the presence of the hunger signal, cyclic AMP (cAMP), CAP (catabolite activator protein) protein (activator) binds to the promote to enhance the transcription of the lacZ gene (and others on the operon) – Fig. 10-13 b T. cAMP-CAP and RNA polymerase mutually stimulate the binding of themselves to the lac control region via cooperative binding. 4. The activity of adenylate cyclase (which catabolizes the production of cAMP from ATP) is down-regulated by glucose – Fig. 10-13a. B. The structures of target DNA sites U. The CAP-cAMP binding to a site different from that for the lacI repressor, although both bind to the 5’ end of the operon – Fig. 10-14. V. The bending of DNA, which is promoted by the CAP-cAMP complex, may aid the binding of RNA polymerase to the promoter. W. CAP and DNA polymerase bind directly adjacent to each other on the lac promoter – Fig. 10-16. X. A summary of the lac operon – Fig. 10-17. D. Comparison of repression and activation – Fig. 10-18. V. Dual positive and negative control: the arabinose operon. A. The structure of the ara operon – Fig.10-19 B. Regulation: • In the presence of arabinose, the AraC protein binds to the araI region (the initiator region, containing both an operator site and a promoter) and the CAP-cAMP complex binds to a site adjacent to araI, leading to the expression of the araB, araA and araD genes (three structural genes that encode the metabolic enzymes to break down the sugar arabinose). 2 Bio300 Handout #15 • Fall, 2004 In the absence of arabinose, the AraC protein functions as a repressor by binding to both the araI and araO regions, forming a DNA loop. This binding prevents transcription of the ara operon. Fig. 10-20. 3