* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Presentation
Cre-Lox recombination wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Genome evolution wikipedia , lookup
Epigenetics wikipedia , lookup
DNA vaccination wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Genomic imprinting wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Epitranscriptome wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Oncogenomics wikipedia , lookup
Genome (book) wikipedia , lookup
Ridge (biology) wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Non-coding RNA wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Microevolution wikipedia , lookup
Non-coding DNA wikipedia , lookup
Epigenomics wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Minimal genome wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Designer baby wikipedia , lookup
Cancer epigenetics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Gene expression profiling wikipedia , lookup
Point mutation wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Primary transcript wikipedia , lookup
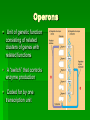
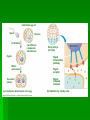
Gene Expression Cell Differentiation Cell types are different because genes are expressed differently in them. Causes: Changes in chromatin structure Initiation of transcription RNA processing mRNA degradation Translation Protein processing and degradation Operons Unit of genetic function consisting of related clusters of genes with related functions A “switch” that controls enzyme production Coded for by one transcription unit Repressible Operons (The trp Operon) Repressible Operon: always operates unless a repressor turns it off. promoter: RNA polymerase binding site; begins transcription operator: controls access of RNA polymerase to genes transcription stops here when repressor is in place repressor: protein that binds to operator and prevents attachment of RNA polymerase Sometimes, a corepressor must be in place for the repressor to be active Tryptophan (a.a.) synthesis Transcription is repressed when tryptophan binds to the repressor, which connects to the operator Inducible Operons (The lac operon) Inducible Operon: Always off unless an inducer is present Inducer attaches to the repressor and causes it to move so that transcription can occur Lactose metabolism (lac operon) lactose not present: repressor active, operon off; no transcription for lactose enzymes lactose present: repressor inactive, operon on inducer molecule inactivates protein repressor (allolactose) Chromatin Complex of DNA and proteins DNA Packing histone protein (+ charged amino acids ~ phosphates of DNA are charged) Nucleosome ”beads on a string” basic unit of DNA packing Heterochromatin highly condensed interphase DNA (can not be transcribed) Euchromatin “true chromatin” less compacted interphase DNA (can be transcribed) Histone Modification Genes within highly packed heterochromatin are usually not expressed Chemical modifications to histones and DNA of chromatin influence both chromatin structure and gene expression Acetylation prevents histones from packing tightly, which allows genes to be expressed. Methylation causes histones to pack tightly so that genes are not expressed. Epigenetic Inheritance Expression of traits is not necessarily related to the nucleotide sequence Some individuals may express traits from their genes where others will not based on histone modifications One twin may express a trait or get a disease that the other does not, despite same genes Schizophrenia Some cancers Etc. Regulation of Transcription Control Elementsnoncoding DNA that regulate binding proteins Enhancers- segments that influence how a gene is expressed Often placed far from the actual gene RNA and Protein Processing Alternative RNA splicing Different mRNA molecules formed from the same primary transcript mRNA degradation Protein processing Protein degradation proteasomes Cell Differentiation How cells become specialized in structure and function. Determinants exist in the egg cell Influence the expression of characteristics in different regions of cells Once cells divide by mitosis, specific regions of the embryo will express genes differently Unfertilized egg cell Sperm Fertilization Nucleus Two different cytoplasmic determinants Zygote Mitotic cell division Two-celled embryo (a) Cytoplasmic determinants in the egg Early embryo (32 cells) Signal transduction pathway Signal receptor Signal molecule (inducer) (b) Induction by nearby cells NUCLEUS Body Plan Setup Pattern Formation cytoplasmic determinants inductive signals determine spatial organization of tissues Biology of Cancer Oncogene- cancer-causing genes Proto-oncogene- normal cellular genes How does a proto-oncogene become an oncogene? movement of DNA; chromosome fragments that have rejoined incorrectly amplification; increases the number of copies of proto-oncogenes point mutation; protein product more active or more resistant to degradation Tumor-suppressor genes changes in genes that prevent uncontrolled cell growth (cancer growth stimulated by the absence of suppression) ras and p53 ras Produces Ras proteins Hyperactive Ras protein causes cell cycle to continue (increased cell division) Mutations involved in 30% of all cancers p53 Tumor-suppressor gene Activated by DNA damage Turns on DNA repair or activates “suicide” genes Mutations involved in 50% of all cancers