* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Poliammine, evoluzione e patogenicità in Shigella spp
Copy-number variation wikipedia , lookup
Transposable element wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genetic engineering wikipedia , lookup
Genomic library wikipedia , lookup
Gene therapy wikipedia , lookup
Oncogenomics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Essential gene wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Gene nomenclature wikipedia , lookup
Gene desert wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Public health genomics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Ridge (biology) wikipedia , lookup
X-inactivation wikipedia , lookup
Genomic imprinting wikipedia , lookup
History of genetic engineering wikipedia , lookup
Minimal genome wikipedia , lookup
Epigenetics of human development wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression programming wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Microevolution wikipedia , lookup
Designer baby wikipedia , lookup
Genome evolution wikipedia , lookup
Gene expression profiling wikipedia , lookup
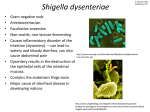
Polyamine, evolution and pathogenicity in Shigella spp Barbagallo M1, Di Martino ML1, Marcocci L2, Pietrangeli P2, De Carolis E3, Colonna B1, Prosseda G1 Dip Biologia e Biotecnologie “C. Darwin” Università di Roma “Sapienza” Dip di Scienze Biochimiche “A. Rossi Fanelli”, Università di Roma “Sapienza” 3 Istituto di Microbiologia, Università Cattolica del Sacro Cuore Rome, Italy 1 2 Shigella, the causative agent of bacillary dysentery, is able to cross the epithelial lining, to induce apoptotic killing of macrophages, and to enter and spread into epithelial cells, eliciting the inflammatory destruction of the intestinal epithelial barrier. These processes require coordinated expression of virulence genes residing on a large plasmid and on the chromosome. The genomes of Shigella and E. coli, its commensal ancenstor, are colinear and highly homologous. Critical events in the evolution of Shigella have been the acquisition of the virulence plasmid through lateral gene transfer and the inactivation of genes affecting the full expression of the virulence phenotype. In this context we have analyzed to which extent the presence of the plasmid-encoded virF gene, the major activator of the Shigella invasivity regulon, has modified the transcriptional profile of E. coli. Combining results from transcriptome assays and comparative genome analyses we show that in E.coli VirF, besides being able to up-regulate several chromosomal genes potentially able to increase bacterial fitness within the host, also activates genes which have been lost by Shigella. We have focused our attention on the speG gene, which encodes spermidine acetyltransferase, an enzyme catalyzing the conversion of spermidine into the physiological inert acetylspermidine, since recent evidence stresses the involvement of polyamines in bacterial pathogenesis. We show that the speG gene is inactive in Shigella and that the consequent accumulation of spermidine strongly favours the survival of the pathogen under oxidative stress conditions, as well as within the macrophages it invades during infection. Our data uncover an as yet unknown aspect of the pathogenicity strategy adopted by Shigella and contribute to decipher the complexity of the invasivity process.