* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Model of Skeletal Muscle Energy Metabolism
Multi-state modeling of biomolecules wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Metalloprotein wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Epitranscriptome wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Photosynthesis wikipedia , lookup
Mitochondrion wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Biosynthesis wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Lactate dehydrogenase wikipedia , lookup
Electron transport chain wikipedia , lookup
Microbial metabolism wikipedia , lookup
Biochemistry wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
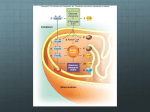
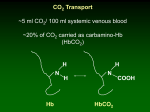
MATERIALS S2: METABOLIC REACTIONS FLUX EXPRESSIONS The flux expressions for the compartmentalized lumped metabolic reactions that convert substrates to products in the two subcellular compartments (cytosol and mitochondria) in coupled with the energy controller pairs ATP-ADP and NADH-NAD+ are written here from the generalized reaction flux expression (equation (5) of the manuscript) which is based on a phenomenological one-step Michaelis-Menten kinetics for enzymatic reactions. These expressions differ from those of our previous model [1] through their dependencies on the compartmentalized (cytosolic or mitochondrial) metabolites concentrations, including the compartmentalized energy controller ratios ATP/ADP and NADH/NAD+. As necessary, further descriptions are given below for the individual reactions and the reaction flux expressions. Reactions in Cytosol 1. Glucose Utilization GLCG6P GLC ATP G6P ADP V GLCG6P 1 Ccyt,G6P G6P KGLC G6P Ccyt,ATP Ccyt,ADP Ccyt,ATP K PS+ GLC G6P Ccyt,ADP Ccyt,GLC KGLCG6P Ccyt,GLC Ccyt,G6P G6P 1 K KGLCG6P GLCG6P This reaction is catalyzed by the enzyme hexokinase which is inhibited by G6P ([5], Ch. 15; [6], Ch. 19). The exact inhibition mechanism in-vivo is not well known. Therefore, a combination of noncompetitive and product inhibition mechanism ([7], Ch. 3) is considered, which effectively modifies the Vmax as well as the Km of the reaction. 2. Glycogen Synthesis G6PGLY G6P ATP GLY ADP 2 PI Ccyt,ATP Ccyt,G6P Ccyt,ADP KG6PGLY VG6PGLY Ccyt,ATP Ccyt,G6P PS+ KG6PGLY C 1 K cyt,ADP G6PGLY This is a sum of 4 enzymatic reactions G6P G1P, G1P+UTP UDP-GLC + 2 PI, UDP-GLC + GLYn UDP + GLYn+1 and UDP+ATP UTP+ADP catalyzed by the enzymes phosphoglucomutase, UDPglucose pyrophosphorylase, glycogen synthase, and nucleoside diphosphokinase ([5], Ch. 20; [6], Ch. 23). 3. Glycogen Breakdown GLYG6P GLY PI G6P Ccyt,AMP Ccyt,GLYCcyt,PI Ccyt,ATP KGLYG6P VGLYG6P Ccyt,AMP Ccyt,GLYCcyt,PI PS KGLYG6P C 1 K cyt,ATP GLY G6P This is a sum of 2 enzymatic reactions GLY + PI G1P and G1P G6P catalyzed by the enzymes glycogen phosphorylase and phosphoglucomutase ([5], Ch. 20; [6], Ch. 23). The activity of glycogen phosphorylase is regulated by AMP and ATP; AMP acts as a positive effector (activator) and ATP acts a negative effector (inhibitor) by competing with AMP. So the reaction is controlled by CAMP/CATP ratio. G6P ATP 2 GA3P ADP Ccyt,ATP Ccyt,G6P Ccyt,ADP KG6PGA3P VG6PGA3P Ccyt,ATP Ccyt,G6P PS+ KG6PGA3P C 1 K cyt,ADP G6PGA3P 4. Glucose 6-Phosphate Breakdown G6PGA3P This is a sum of 4 enzymatic reactions G6P F6P, F6P + ATP F16BP + ADP, F16BP DHAP + GA3P, and DHAP GA3P catalyzed by the enzymes phosphoglucose isomerase, phosphofructokinase, aldolase, and triose phosphate isomerase ([5], Ch. 15; [6], Ch. 19). GA3P PI NAD 13BPG NADH 5. Glyceraldehyde 3-Phosphate Breakdown GA3P13BPG Ccyt,NAD+ Ccyt,NADH VGA3P13BPG RS Ccyt,NAD+ K GA3P13BPG Ccyt,NADH Ccyt,GA3PCcyt,PI K GA3P13BPG Ccyt,GA3PCcyt,PI 1 K GA3P13BPG This reaction is catalyzed by the enzyme glyceraldehyde 3-phosphate dehydrogenase and is known to be reversible ([5], Ch. 15; [6], Ch. 19). Since |G| of the reaction is non-zero, we consider it as irreversible. 6. Pyruvate Production 13BPG PYR 13BPG 2 ADP PYR 2 ATP Ccyt,ADP Ccyt,13BPG Ccyt,ATP K13BPG PYR V13BPG PYR Ccyt,ADP Ccyt,13BPG PS K13BPG PYR 1 Ccyt,ATP K13BPG PYR This is a sum of 4 enzymatic reactions 13BPG + ADP 3PG + ATP, 3PG 2PG, 2PG PEP, and PEP + ADP PYR+ATP catalyzed by the enzymes phosphoglycerate kinase, phosphoglycerate mutase, enolase, and pyruvate kinase ([5], Ch. 15; [6], Ch. 19). 7. Pyruvate Reduction PYR LAC PYR NADH LAC NAD Ccyt,NADH Ccyt,PYR Ccyt,NAD+ K PYR LAC VPYR LAC RS+ Ccyt,NADH Ccyt,PYR K PYR LAC 1 Ccyt,NAD+ K PYR LAC This is an important reaction in skeletal muscle metabolism. When oxygen availability is limited (e.g., during muscle ischemia or intense muscle activities), NADH in cytosol can accumulate and reduce pyruvate to lactate with the help of the enzyme lactate dehydrogenase ([5], Ch. 15; [6], Ch. 19). 1 LAC NAD PYR NADH Ccyt,NAD+ Ccyt,LAC Ccyt,NADH K LAC PYR LACPYR VLACPYR RSCcyt,NAD+ Ccyt,LAC K LAC PYR 1 K LAC PYR Ccyt,NADH This is the reverse lactate dehydrogenase reaction. During aerobic metabolism, NAD+ in cytosol can increase and oxidize lactate to pyruvate. 8. Lactate Oxidation 9. Alanine Production PYR ALA PYR ALA Ccyt,PYR K PYR ALA VPYR ALA Ccyt,PYR 1 K PYR ALA GA3P 3 FAC NADH TGL 3 CoA PI NAD Ccyt,NADH Ccyt,GA3P Ccyt,FAC Ccyt,NAD+ K GA3P TGL VGA3P TGL RS+ Ccyt,NADH Ccyt,GA3P Ccyt,FAC K GA3P TGL 1 Ccyt,NAD+ K GA3P TGL 10. Triglyceride Synthesis GA3P TGL This is a sum of several enzymatic reactions. It can be viewed as lumping of reactions GA3P DHAP, DHAP + NADH G3P + NAD+, and G3P + 3 FAC TGL + 3 CoA + PI. The major enzymes are glycerol-3 phosphate dehydrogenase and acyltransferase ([5], Ch. 21; [6], Ch. 24). The synthesis of triglycerides from glycerol is neglected here as the activity of the enzyme glycerol kinase is negligible in muscle. 11. Lipolysis (Triglycerides Hydrolysis) TGLFFA 12. Fatty Acyl-CoA Formation FFA FAC TGL GLR 3 FFA Ccyt,TGL K TGLFFA VTGL FFA Ccyt,TGL 1 K TGL FFA FFA CoA 2 ATP FAC 2 ADP 2 PI Ccyt,ATP Ccyt,ADP VFFA FAC Ccyt,ATP PS+ K FFA FAC Ccyt,ADP Ccyt,FFA Ccyt,CoA K FFA FAC Ccyt,FFA Ccyt,CoA 1 K FFA FAC This reaction is also called fatty acid activation in which the enzyme is fatty acid thiokinase. The activated fatty acid is transported into the mitochondrial matrix through the carnitine shuttle which is subsequently oxidized to ACoA by several enzymatic reactions ([5], Ch.21; [6], Ch.24). ATP ADP PI 13. ATP Hydrolysis 2 ATPADP V ATPADP 1 Ccyt,PI PI K ATP ADP Ccyt,ATP Ccyt,ADP Ccyt,ATP K PS+ ATP ADP Ccyt,ADP This reaction is catalyzed by the enzyme ATPase and is the primary source of energy supply for muscle contraction ([5], Ch.14; [6], Ch.17). This reaction is inhibited by ADP and PI. PCR ADP CR ATP Ccyt,ADP Ccyt,PCR Ccyt,ATP K PCR CR VPCR CR Ccyt,ADP Ccyt,PCR PS K PCR CR C 1 K cyt,ATP PCR CR 14. Phosphocreatine Breakdown PCR CR This is an ATP buffer reaction catalyzed by the enzyme creatine kinase. It functions to maintain ATP homeostasis during muscle contraction ([5], Ch.14; [6], Ch.17). It is the primary source of immediate energy supply during the transitions from rest to exercise. CR ATP PCR ADP 15. Phosphocreatine Synthesis CR PCR Ccyt,ATP Ccyt,ADP VCR PCR Ccyt,ATP PS+ KCR PCR C cyt,ADP Ccyt,CR KCR PCR Ccyt,CR 1 K CR PCR This is the reverse creatine kinase reaction where creatine is phosphorylated to phosphocreatine. AMP ATP ADP ADP 16. Adenylate Kinase – Forward AMPADP Ccyt,ATP Ccyt,ADP VAMPADP Ccyt,ATP PS+ K AMP ADP Ccyt,ADP Ccyt,AMP K AMPADP Ccyt,AMP 1 K AMPADP ADP ADP AMP ATP Ccyt,ADP Ccyt,ADP Ccyt,ATP K ADPAMP VADPAMP Ccyt,ADP Ccyt,ADP PSK 1 ADP AMP Ccyt,ATP K ADPAMP 17. Adenylate Kinase – Reverse ADPAMP Reactions in Mitochondria 18. Pyruvate Oxidation PYR CoA NAD ACoA NADH CO 2 3 PYR ACoA Cmit,NAD+ Cmit,NADH VPYR ACoA Cmit,NAD+ RS K PYR ACoA Cmit,NADH Cmit,PYR Cmit,CoA K PYR ACoA Cmit,PYR Cmit,CoA 1 K PYR ACoA This is the first reaction inside the mitochondrial matrix in which ACoA is formed from the oxidative decarboxylation of pyruvate (carbohydrate oxidation) by the enzyme pyruvate dehydrogenase leading to the TCA cycle ([5], Ch.16; [6], Ch.20). FAC 7 CoA 14 NAD 8 ACoA 14 NADH Cmit,NAD+ Cmit,FACCmit,CoA Cmit,NADH K FACACoA VFACACoA Cmit,NAD+ Cmit,FACCmit,CoA RS K FACACoA 1 K C FAC ACoA mit,NADH 19. Fatty Acyl-CoA Oxidation FACACoA This reaction producing ACoA from the activated fatty acid (fat) inside the mitochondrial matrix is highly complex. It is the result of combining 7 cycles of reactions in which each cycle consists of 4 enzymatic reactions catalyzed by the enzymes acyl-CoA dehydrogenase, enoyl-CoA hydratase, beta-hydroxyacylCoA dehydrogenase, and acyl-CoA acetyletransferase (thiolase) ([5], Ch. 21; [6], Ch. 24). For simplicity, the reducing equivalents FAD and FADH2 are considered equivalent to NAD+ and NADH, as they consume equal amount of O2. ACoA OXA CIT CoA 20. Citrate Production ACoACIT Cmit,ACoACmit,OXA K ACoACIT VACoACIT 1 Cmit,ACoACmit,OXA K ACoACIT This is the first reaction of TCA cycle catalyzed by the enzyme citrate synthase ([5], Ch. 16; [6], Ch. 20). CIT NAD AKG NADH CO 2 21. Alpha-Ketoglutarate Production CITAKG Cmit,NAD+ Cmit,NADH VCIT AKG Cmit,NAD+ RS K CIT AKG Cmit,NADH Cmit,CIT K CITAKG Cmit,CIT 1 K CIT AKG This is a sum of two enzymatic reactions CIT ↔ ICIT and ICIT+NAD+ → AKG+CO2+NADH catalyzed by the enzymes aconitase and isocitrate dehydrogenase. 22. Succinyl-CoA Production AKG SCoA AKG CoA NAD SCoA NADH CO2 Cmit,NAD+ Cmit,NADH VAKG SCoA RS C + K AKG SCoA mit,NAD Cmit,NADH 4 Cmit,AKG Cmit,CoA K AKG SCoA Cmit,AKG Cmit,CoA 1 K AKG SCoA 23. Succinate Production SCoA SUC SCoA GDP PI SUC CoA GTP Cmit,ADP Cmit,SCoA Cmit,PI Cmit,ATP KSCoA SUC VSCoA SUC Cmit,ADP Cmit,SCoA Cmit,PI PS KSCoA SUC C 1 K mit,ATP SCoA SUC Because the reaction GTP+ADP GDP+ATP is in fast equilibrium, we assume the GTP/GDP ratio proportional to the ATP/ADP ratio ([5], Ch. 16; [6], Ch. 20). 24. Malate Production SUCMAL SUC NAD MAL NADH Cmit,NAD+ Cmit,SUC Cmit,NADH KSUCMAL VSUCMAL RS C Cmit,SUC + K MALOXA mit,NAD 1 KSUCMAL Cmit,NADH This is a sum of two enzymatic reactions SUC + FAD → FUM + FADH2 & FUM ↔ MAL catalyzed by the enzymes succinate dehydrogenase and fumarate ([5], Ch.16; [6], Ch.20); FAD and FADH2 are considered equivalent to NAD+ and NADH, as they consume equal amount of O2. MAL NAD OXA NADH 25. Oxaloacetate Production MALOXA 26. Oxygen Utilization O2H2O Cmit,NAD+ Cmit,NADH VMALOXA RS C + K MALOXA mit,NAD Cmit,NADH Cmit,MAL K MALOXA Cmit,MAL 1 K MALOXA O2 6 ADP 6 PI 2 NADH 2 H 2O 6 ATP 2 NAD Cmit,ADP Cmit,ATP VO2H2O Cmit,ADP PS K O2H2O C mit,ATP Cmit,NADH Cmit,NAD+ Cmit,NADH RS K O2H2O C mit,NAD + F Cmit,O2 Cmit,PI K O2H2O F Cmit,O2 Cmit,PI 1 K O2H2O This is also the ATP synthesis reaction. This is a sum of several enzymatic reactions at complex I–V that constitute the electron transport chain and oxidative phosphorylation inside the mitochondrial matrix ([5], Ch.19; [6], Ch.21); FAD and FADH2 are considered equivalent to NAD+ and NADH as they consume equal amount of O2. Furthermore, 1 NADH is assumed to produce 3 ATP (i.e., P/O ratio = 3; a perfect coupling with negligible proton leak). 5 Reference List 1. Dash RK, Li Y, Kim J, Saidel GM, Cabrera ME (2008) Modeling cellular metabolism and energetics in skeletal muscle: large-scale parameter estimation and sensitivity analysis. IEEE Trans Biomed Eng 55: 1298-1318. 2. Dash RK, Bassingthwaighte JB (2004) Blood HbO2 and HbCO2 dissociation curves at varied O2, CO2, pH, 2,3-DPG and temperature levels. Ann Biomed Eng 32: 1676-1693. 3. Dash RK, Bassingthwaighte JB (2006) Simultaneous blood-tissue exchange of oxygen, carbon dioxide, bicarbonate, and hydrogen ion. Ann Biomed Eng 34: 1129-1148. 4. Geers C, Gros G (2000) Carbon dioxide transport and carbonic anhydrase in blood and muscle. Physiol Rev 80: 681-715. 5. Nelson D, Cox M (2000) Lehninger Principles of Biochemistry (third edition). Worth Publishers, New York. 6. Stryer L (1996) Biochemistry (fourth edition). W.H. Freeman and Company, New York. 7. Segel I (1993) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. Wiley-Interscience, New York. 6