* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download chapt06HOv2.ppt
Fatty acid synthesis wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Biosynthesis wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Photosynthesis wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Glyceroneogenesis wikipedia , lookup
Phosphorylation wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Microbial metabolism wikipedia , lookup
Biochemistry wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Electron transport chain wikipedia , lookup
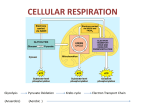
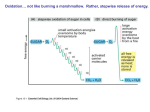
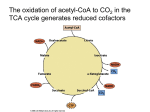
Microbial Metabolism § All cells need to accomplish two fundamental tasks • Synthesize new parts • Cell walls, membranes, ribosomes, nucleic acids • Harvest energy to power reactions • Sum total of these is called metabolism • Human implications • • • • • Used to make biofuels Used to produce food Important in laboratory Invaluable models for study Unique pathways potential drug targets 6.1. Principles of Metabolism Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. § Can separate metabolism into two parts CATABOLISM ANABOLISM Energy source (glucose) Cell structures (cell wall, membrane, ribosomes, surface structures) • Catabolism • Processes that degrade compounds to release energy • Cells capture to make ATP Energy Macromolecules (proteins, nucleic acids, polysaccharides, lipids) Energy • Anabolism Subunits (amino acids, nucleotides, sugars, fatty acids) • Biosynthetic processes • Assemble subunits of macromolecules • Use ATP to drive reactions • Processes intimately linked Energy Precursor metabolites Waste products Nutrients (acids, carbon dioxide) (source of nitrogen, sulfur, etc.) Catabolic processes harvest the energy released during the breakdown of compounds and use it to make ATP. The processes also produce precursor metabolites used in biosynthesis. Anabolic processes (biosynthesis) synthesize and assemble subunits of macromolecules that make up the cell structures. The processes use the ATP and precursor metabolites produced in catabolism. Harvesting Energy § Free energy is energy available to do work • E.g., energy released when chemical bond is broken • Compare free energy of reactants, products • Exergonic reactions: reactants have more free energy • Energy is released in reaction • Endergonic reactions: products have more free energy • Reaction requires input of energy • Change in free energy is same regardless of number of steps involved (e.g., converting glucose to CO2 + H2O) • Cells use multiple steps when degrading compounds • Energy released from exergonic reactions powers endergonic reactions 1 Fig. 6.4 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Intermediatea Starting compound Intermediateb End product Intermediateb1 End product1 Intermediateb2 End product2 (a) Linear metabolic pathway Starting compound Intermediatea (b) Branched metabolic pathway Starting compound Intermediated End product Intermediatea Intermediatec Intermediateb (c) Cyclical metabolic pathway Fig. 6.9 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Glucose molecules To: Lipid synthesis To: Amino acid synthesis To: Carbohydrate synthesis To: Nucleic acid synthesis CO2 molecules + energy Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Terminal electron acceptors Energy sources Energy released Organic carbon compounds Organic carbon compounds H 2S CO2 SO4 S0 FeOOH Fe2+ NH4+ NO2– ( to form NH4+) NO3– ( to form NH4+) Mn2+ MnO2 Relative tendency to give up electrons Relative tendency to give up electrons H2 NO3– ( to form NH2) O2 (a) Energy is released when electrons are moved from an energy source with a low affinity for electrons to a terminal electron acceptor with a higher affinity. Terminal electron acceptors Inorganic energy sources H2 Pyruvate NO3– (to form NH4+) O2 (b) Three examples of chemoorganotrophic metabolism Relative tendency to give up electrons Glucose Terminal electron acceptors H 2S CO2 Relative tendency to give up electrons Glucose as an energy source Relative tendency to give up electrons Fig. 6.7 Fe2+ O2 (c) Three examples of chemolithotrophic metabolism 2 Components of Metabolic Pathways § Role of the Chemical Energy Source and the Terminal Electron Acceptor § Some atoms, molecules more electronegative than others Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Energy release Organic carbon compounds H2 Organic carbon compounds H 2S CO2 SO4 S0 FeOOH Fe2+ NH4+ NO2– ( to form NH4+) NO3– ( to form NH4+) Mn2+ MnO2 Relative tendency to give up electrons • (E.g., glucose to O2) Terminal electron acceptors Energy sources Relative tendency to give up electrons • Greater affinity for electrons • Energy released when electrons move from low affinity molecule to high affinity molecule NO3– ( to form NH2) O2 (a) Energy is released when electrons are moved from an energy source with a low affinity for electrons to a terminal electron acceptor with a higher affinity. Components of Metabolic Pathways § Role of the Chemical Energy Source and the Terminal Electron Acceptor (continued…) § More energy released when difference in electronegativity is greater Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. • Acceptor: • Terminal electron acceptor Glucose Pyruvate NO3– (to form NH4–) O2 (b) Three examples of chemoorganotrophic metabolism Terminal electron acceptors H2 H 2S CO2 Fe2+ Relative tendency to give up electrons • Energy source Inorganic energy sources Relative tendency to give up electrons • Electron donor: Relative tendency to give up electrons Glucose Terminal as an electron energy source acceptors O2 (c) Three examples of chemolithotrophic metabolism Components of Metabolic Pathways § Role of Electron Carriers • Energy harvested in stepwise process • Electrons transferred to electron carriers, which represent reducing power (easily transfer electrons to molecules) – Raise energy level of recipient molecule • NAD+/NADH, NADP+/NADPH, and FAD/FADH2 3 Components of Metabolic Pathways § Role of ATP • Adenosine triphospate (ATP) is energy currency • • • • Composed of ribose, adenine, three phosphate groups Adenosine diphospate (ADP) acceptor of free energy Cells produce ATP by adding Pi to ADP using energy Release energy from ATP to yield ADP and Pi § Three processes to generate ATP • Substrate-level phosphorylation Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Unstable (high-energy) bonds • Exergonic reaction powers • Oxidative phosphorylation • Proton motive force drives • Photophosphorylation • Sunlight used to create proton motive force to drive P ~ P~ P ATP Pi Pi Energy used The energy comes from catabolic reactions. Energy released The energy drives anabolic reactions. P~ P ADP 6.3. The Central Metabolic Pathways § ATP § Reducing power: NADH, FADH2, NADPH § Precursor metabolites • Glucose molecules can have different fates • Can be completely oxidized to CO2 for maximum ATP • Can be siphoned off as precursor metabolite for use in biosynthesis Precursor Metabolites § Precursor metabolites are intermediates of catabolism that can be used in anabolism • Serve as carbon skeletons for building macromolecules • E.g., pyruvate can be converted into amino acids alanine, leucine, or valine 4 Components of Metabolic Pathways § Prokaryotes remarkably diverse in using energy sources and terminal electron acceptors • Organic, inorganic compounds used as energy source • O2, other molecules used as terminal electron acceptor • Electrons removed through series of oxidation-reduction reactions or redox reactions • Substance that loses electrons is oxidized • Substance that gains electrons is reduced • Electron-proton pair, or Transfer of electrons hydrogen, actually moves e • Dehydrogenation = oxidation Compound + Compound X Y • Hydrogenation = reduction X loses electron(s). Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. – e– Compound X + Compound Y (reduced) (oxidized) Y gains electron(s). X is the reducing agent. Y is the oxidizing agent. X is oxidized by the reaction. Y is reduced by the reaction. Overview of Catabolism § Three central metabolic pathways • Oxidize glucose to CO2 • Catabolic, but precursor metabolites and reducing power can be diverted for use in biosynthesis • Termed amphibolic to reflect dual role • Glycolysis • Splits glucose (6C) to two pyruvates (3C) • Generates modest ATP, reducing power, precursors • Pentose phosphate pathway • Primary role is production precursor metabolites, NADPH • Tricarboxylic acid cycle • Oxidizes pyruvates from glycolysis • Generates reducing power, precursor metabolites, ATP Overview of Catabolism Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. GLUCOSE § Central metabolic pathways • Glycolysis • Pentose phosphate pathway • Tricarboxylic acid cycle § Key outcomes • ATP • Reducing power • Precursor metabolites 2 Pentose phosphate pathway Starts the oxidation of glucose Glycolysis Oxidizes glucose to pyruvate 1 Yields ~ ~ + Reducing power ATP by substrate-level phosphorylation Yields Reducing power Biosynthesis 5 3a Fermentation Reduces pyruvate or a derivative Acids, alcohols, and gases Pyruvate Pyruvate Transition step CO2 CO2 Yields Reducing power AcetylCoA AcetylCoA X2 CO2 CO2 3b TCA cycle Incorporates an acetyl group and releases CO2 (TCA cycles twice) Yields ATP by substrate-level phosphorylation ~ ~ + Reducing power 4 Respiration Uses the electron transport chain to convert reducing power to proton motive force Yields ~ ~ ATP by oxidative phosphorylation 5 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Glycolysis Pentose phosphate pathway Glucose 6-phosphate Lipopolysaccharide (polysaccharide) Fructose 6-phosphate Ribose 5-phosphate Nucleotides amino acids (histidine) Erythrose 5-phosphate Peptidoglycan Dihydroxyacetone phosphate Amino acids (phenylalanine, tryptophan, tyrosine) Lipids (glycerol component) 3-phosphoglycerate Amino acids (cysteine, glycine, serine) Phosphoenolpyruvate Amino acids (phenylalanine, tryptophan, tyrosine) Anabolic Pathways— Synthesizing Subunits from Precursor Molecules Pyruvate Pyruvate Acetyl-CoA Acetyl-CoA Amino acids (alanine, leucine, valine) Lipids (fatty acids) Oxaloacetate Amino acids (aspartate, asparagine, isoleucine, lysine, methionine, threonine) α- ketoglutarate X2 Amino acids (arginine, glutamate, glutamine, proline) TCA cycle 6.3. The Central Metabolic Pathways § Pentose Phosphate Pathway • Also breaks down glucose • Important in biosynthesis of precursor metabolites • Ribose 5-phosphate, erythrose 4-phosphate • Also generates reducing power: NADPH • Yields vary depending upon alternative taken Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. GLUCOSE 2 Pentose phosphate pathway Starts the oxidation of glucose Glycolysis Oxidizes glucose to pyruvate 1 Yields ~ ~ + Reducing power ATP by substrate-level phosphorylation Yields Reducing power Biosynthesis 5 3a Fermentation Reduces pyruvate or a derivative Acids, alcohols, and gases Pyruvate Pyruvate Transition step CO2 CO2 Yields Reducing power AcetylCoA AcetylCoA X2 CO2 CO2 3b TCA cycle Incorporates an acetyl group and releases CO2 (TCA cycles twice) Yields ~ ATP by substrate-level phosphorylation ~ + Reducing power 4 Respiration Uses the electron transport chain to convert reducing power to proton motive force Yields ~ ~ ATP by oxidative phosphorylation 6 6.3. The Central Metabolic Pathways Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. GLUCOSE Pentose phosphate pathway Starts the oxidation of glucose 2 Yields Glycolysis Oxidizes glucose to pyruvate 1 P~ P~ P + Reducing power ATP by substrate-level phosphorylation § Glycolysis Yields Reducing power Biosynthesis 5 Fermentation Reduces pyruvate or a derivative Acids, alcohols, and gases 3a Glucose Transition step CO2 CO2 Yields • Converts 1 glucose to 2 pyruvates; yields net 2 ATP, 2 NADH • Investment phase: Reducing power Pyruvate Pyruvate x2 CO2 TCA cycle Incorporates an acetyl group and releases CO2 (TCA cycles twice) Yields P~ P ~ P + ~ ADP Reducing power ATP by substrate-level phosphorylation 1 ATP is expended to add a phosphate group. ~ ~ ATP CO2 3b Respiration Uses the electron transport Chain to convert reducing power to proton motive force 4 Yields Glucose 6-phosphate P~ P~ P ATP by oxidative phosphorylation 2 A chemical rearrangement occurs. Fructose 6-phosphate ~ ~ ATP ATP is expended to add a phosphate group. 4 The 6-carbon molecule is split into two 3-carbon molecules. ~ ADP • 2 phosphate groups added • Glucose split to two 3-carbon molecules 3 Fructose 1,6-bisphosphate Dihydroxyacetone phosphate A chemical rearrangement of one of the molecules occurs. 5 Glyceraldehyde 3-phosphate NAD+ NAD+ NADH + H+ 1,3-bisphosphoglycerate • Pay-off phase: ADP ~ ~ The addition of a phosphate group is coupled to a redox reaction, generating NADH and a high-energy phosphate bond. ~ ~ ATP • 3-carbon molecules converted to pyruvate • Generates 4 ATP, 2 NADH total 6 NADH + H+ ATP is produced by substrate-level phosphorylation. 7 ~ ~ ~ ~ 3-phosphoglycerate 8 2-phosphoglycerate A chemical rearrangement occurs. 9 H 2O H 2O Phosphoenolpyruvate ADP ~ ~ ATP Water is removed, causing the phosphate bond to become high-energy. 10 ATP is produced by substrate-level phosphorylation. ~ ~ ~ ~ Pyruvate 6.3. The Central Metabolic Pathways § Transition Step Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. GLUCOSE • CO2 is removed from pyruvate • Electrons reduce NAD+ to NADH + H+ • 2-carbon acetyl group joined to coenzyme A to form acetyl-CoA • Takes place in mitochondria in eukaryotes 2 Pentose phosphate pathway Starts the oxidation of glucose Yields Glycolysis Oxidizes glucose to pyruvate 1 ~ ~ + Reducing power ATP by substrate-level phosphorylation Pyruvate Yields CO2 Reducing power Biosynthesis Pyruvate CO2 NAD+ Acids, alcohols, and gases CoA Transition step Yields Transition step: CO2 is removed, a redox reaction generates NADH, and coenzyme A is added. Fermentation Reduces pyruvate or a derivative 5 Pyruvate 3a CO2 Reducing power AcetylCoA AcetylCoA NADH + H+ x2 CO2 CoA CO2 TCA cycle Incorporates an acetyl group and releases CO2 (TCA cycles twice) 3b Acetyl-CoA 1 The acetyl group is transferred to oxaloacetate to start a new round of the cycle. Yields ~ ~ + Reducing power ATP by substrate-level phosphorylation CoA Respiration Uses the electron transport chain to convert reducing power to proton motive force 4 Yields ~ ~ ATP by oxidative phosphorylation NADH + H+ A redox reaction generates NADH. 8 Oxaloacetate 2 A chemical rearrangement occurs. Citrate NAD+ Isocitrate NAD+ Malate Water is added. 7 3 A redox reaction generates NADH and CO2 is removed. NADH + H+ H 2O CO2 Fumarate α-ketoglutarate NAD+ 4 FADH2 6 CoA A redox reaction generates FADH2- NADH + H+ FAD 5 The energy released during CoA removal is harvested to produce ATP. CoA Succinyl-CoA Succinate CoA A redox reaction generates NADH, CO2 is removed, and coenzyme A is added. CO2 ~ + Pi ~ ~ ATP ADP 6.3. The Central Metabolic Pathways § Tricarboxylic Acid (TCA) Cycle Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. GLUCOSE 2 Pentose phosphate pathway Starts the oxidation of glucose 2 CO2 2 ATP 6 NADH 2 FADH2 Precursor metabolites ~ + Reducing power Pyruvate CO2 Reducing power Biosynthesis 3a Pyruvate CO2 NAD+ Acids, alcohols, and gases CoA Transition step Yields Transition step: CO2 is removed, a redox reaction generates NADH, and coenzyme A is added. Fermentation Reduces pyruvate or a derivative 5 Pyruvate • Completes oxidation of glucose • • • • • ~ ATP by substrate-level phosphorylation Yields CO2 Reducing power AcetylCoA AcetylCoA § Produces Yields Glycolysis Oxidizes glucose to pyruvate 1 NADH + H+ x2 CO2 CoA CO2 3b TCA cycle Incorporates an acetyl group and releases CO2 (TCA cycles twice) Acetyl-CoA 1 The acetyl group is transferred to oxaloacetate to start a new round of the cycle. Yields ~ ~ + Reducing power ATP by substrate-level phosphorylation CoA Respiration Uses the electron transport chain to convert reducing power to proton motive force 4 Yields ~ ~ ATP by oxidative phosphorylation A redox reaction generates NADH. 8 NADH + H+ Oxaloacetate 2 A chemical rearrangement occurs. Citrate NAD+ Isocitrate NAD+ Malate Water is added. 7 3 A redox reaction generates NADH and CO2 is removed. NADH + H+ H 2O CO2 Fumarate α-ketoglutarate NAD+ 4 FADH2 6 CoA A redox reaction generates FADH2- NADH + H+ FAD 5 The energy released during CoA removal is harvested to produce ATP. CoA Succinyl-CoA Succinate CoA CO2 ~ + Pi ~ ~ ATP A redox reaction generates NADH, CO2 is removed, and coenzyme A is added. ADP 7 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. NH2 NH3 (ammonia) α-ketoglutarate Glutamate is synthesized by adding ammonia to the precursor metabolite α-ketoglutarate. Aspartate NH2 Oxaloacetate Glutamate The amino group (NH2) of glutamate can be transferred to other carbon compounds to produce other amino acids. Fig. 6.30 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. From glycolysis Phenylalanine Branch point II Compound a 3-C Branch point I 7-C compound + Tyrosine 4-C Compound b Tryptophan From pentose phosphate pathway Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. GLUCOSE 2 Pentose phosphate pathway Starts the oxidation of glucose Glycolysis Oxidizes glucose to pyruvate 1 Yields ~ ~ + Reducing power ATP by substrate-level phosphorylation Yields Reducing power Biosynthesis 5 3a Fermentation Reduces pyruvate or a derivative Acids, alcohols, and gases Pyruvate Pyruvate Transition step CO2 CO2 Yields Reducing power AcetylCoA AcetylCoA X2 CO2 CO2 3b TCA cycle Incorporates an acetyl group and releases CO2 (TCA cycles twice) Yields ~ ATP by substrate-level phosphorylation ~ + Reducing power 4 Respiration Uses the electron transport chain to convert reducing power to proton motive force Yields ~ ~ ATP by oxidative phosphorylation 8 Table 6.1 Overview of Catabolism § Respiration transfers electrons from glucose to electron transport chain • Electron transport chain generates proton motive force • Harvested to make ATP via oxidative phosphorylation • Aerobic respiration – O2 is terminal electron acceptor • Anaerobic respiration – Molecule other than O2 as terminal electron acceptor – Also use modified version of TCA cycle 6.4. Respiration § Uses reducing power (NADH, FADH2) generated by glycolysis, transition step, and TCA cycle to synthesize ATP • Electron transport chain generates proton motive force • Drives synthesis of ATP by ATP synthase • Process proposed by British scientist Peter Mitchell in 1961 • Initially widely dismissed • Mitchell conducted years of self-funded research • Received a Nobel Prize in 1978 • Now called chemiosmotic theory 9 Table 6.3 The Electron Transport Chain—Generating Proton Motive Force § Electron transport chain is membrane-embedded electron carriers • • • • • Pass electrons sequentially, eject protons in process Prokaryotes: in cytoplasmic membrane Eukaryotes: in inner mitochondrial membrane Energy gradually released Release coupled to ejection of protons • Creates electrochemical gradient • Used to synthesize ATP • Prokaryotes can also power transporters, flagella Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Electrons from the energy source 2 e– Energy released is used to generate a proton motive force. High energy Low energy Electrons donated to the terminal electron acceptor. 2 1/ 2 H+ O2 H 2O The Electron Transport Chain—Generating Proton Motive Force § Components of an Electron Transport Chain • Most carriers grouped into large protein complexes • Serve as proton pumps • Three general groups are notable • Quinones • Lipid-soluble molecules • Move freely, can transfer electrons between complexes • Cytochromes • Contain heme, molecule with iron atom at center • Several types • Flavoproteins • Proteins to which a flavin is attached • FAD, other flavins synthesized from riboflavin 10 The Electron Transport Chain of Mitochondria Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. GLUCOSE 2 Pentose phosphate pathway Starts the oxidation of glucose Yields Glycolysis Oxidizes glucose to pyruvate 1 P ~ P ~P + Reducing power ATP by substrate-level phosphorylation Yields Reducing power Biosynthesis 5 Pyruvate Pyruvate Eukaryotic cell Fermentation Reduces pyruvate or a derivative Acids, alcohols, and gases 3a Transition step CO2 Yields CO2 Reducing power AcetylCoA AcetylCoA x2 CO2 CO2 3b TCA cycle Incorporates an acetyl group and releases CO2 Inner mitochondrial membrane (TCA cycles twice) Yields Reducing power ATP by substrate-level phosphorylation 4 Respiration Uses the electron transport chain to convert reducing power to proton motive force Yields P P P ATP by oxidative phosphorylation 4 Use of Proton Motive Force Electron Transport Chain Complex III Complex I H+ 4 H+ Ubiquinone 2 H+ 10 1/ 2 2 H+ Intermembrane space Mitochondrial matrix O2 Terminal electron acceptor H 2O NAD+ H+ H+ 2 e– Complex II + ATP synthase (ATP synthesis) Cytochrome c Path of electrons NADH Proton motive force is used to drive: Complex IV 3 ATP + 3 Pi 3 ADP Fig. 6.20 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Prokaryotic cell Cytoplasmic membrane Electron Transport Chain NADH dehydrogenase Uses of Proton Motive Force Ubiquinol veoxidase force rive: H+ (0 or 4) ATP synthase (ATP synthesis) H+ (2 or 4) Ubiquinone Path of electrons 10 Active transport (one mechanism) H+ Rotation of a flagella H+ H+ Proton motive force is used to drive: Transported molecule Outside of cytoplasmic membrane 2 e– – Cytoplasm Succinate dehydrogenase NADH + H+ NAD+ 2 H+ 1/ 2 H 2O O2 Terminal electron acceptor 3 ATP + 3 Pi 3 ADP The Electron Transport Chain—Generating Proton Motive Force § General Mechanisms of Proton Ejection • Some carriers accept only hydrogen atoms (protonelectron pairs), others only electrons • Spatial arrangement in membrane shuttles protons to outside of membrane • When hydrogen carrier accepts electron from electron carrier, it picks up proton from inside cell – or mitochondrial matrix • When hydrogen carrier passes electrons to electron carrier, protons released to outside of cell – or intermembrane space of mitochondria • Net effect is movement of protons across membrane • Establishes concentration gradient • Driven by energy released during electron transfer 11 The Electron Transport Chain—Generating Proton Motive Force § Electron Transport Chain of Mitochondria • Complex I (NADH dehydrogenase complex) • Accepts electrons from NADH, transfers to ubiquinone • Pumps 4 protons • Complex II (succinate dehydrogenase complex) • Accepts electrons from TCA cycle via FADH2, “downstream” of those carried by NADH • Transfers electrons to ubiquinone • Complex III (cytochrome bc1 complex) • Accepts electrons from ubiquinone from Complex I or II • 4 protons pumped; electrons transferred to cytochrome c • Complex IV (cytochrome c oxidase complex) • Accepts electrons from cytochrome c, pumps 2 protons • Terminal oxidoreductase, meaning transfers electrons to terminal electron acceptor (O2) The Electron Transport Chain—Generating Proton Motive Force § Electron Transport Chain of Prokaryotes • Tremendous variation: even single species can have several alternate carriers • E. coli serves as example of versatility of prokaryotes • Aerobic respiration in E. coli • Can use 2 different NADH dehydrogenases – Proton pump equivalent to complex I of mitochondria • Succinate dehydrogenase equivalent to complex II of mitochondria • Can produce several alternatives to optimally use different energy sources, including H2 • Lack equivalents of complex III or cytochrome c – Quinones shuttle electrons directly to ubiquinol oxidase, a terminal oxidoreductase – Two versions for high or low O2 concentrations The Electron Transport Chain—Generating Proton Motive Force § Electron Transport Chain of Prokaryotes (cont…) • Anaerobic respiration in E. coli • Harvests less energy than aerobic respiration – Lower electron affinities of terminal electron acceptors • Some components different • Can synthesize terminal oxidoreductase that uses nitrate as terminal electron acceptor – Produces nitrite – E. coli converts to less toxic ammonia • Sulfate-reducers use sulfate (SO42–) as terminal electron acceptor • Produce hydrogen sulfide as end product 12 The Electron Transport Chain—Generating Proton Motive Force § ATP Synthase—Harvesting the Proton Motive Force to Synthesize ATP • Energy required to establish gradient • Released when gradient is eased • ATP synthase allows protons to flow down gradient in controlled manner • Uses energy to add phosphate group to ADP • 1 ATP formed from entry of ~3 protons • Calculating yields • Based on experiments on rat mitochondria: ~2.5 ATP made per electron pair from NADH ~1.5 ATP made per electron pair from FADH2 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. GLUCOSE Pentose phosphate 2 pathway Starts the oxidation of glucose Yields Glycolysis 1 Oxidizes glucose to pyruvate Yields Reducing power ATP by substrate-level phosphorylation Reducing power Biosynthesis 5 Pyruvate Pyruvate Fermentation Reduces pyruvate or a derivative GLUCOSE Glycolysis Oxidizes glucose to pyruvate ~~ 2 ATP net gain = 0 ~~ 2 ATP Acids, alcohols, and gases 3a Transition step Yield CO2 CO2 Reducing power AcetylCoA AcetylCoA x2 CO2 CO2 TCA cycle 3b Incorporates an acetyl group and releases CO2 (TCA cycles twice) Yields ATP by substrate-level phosphorylation Reducing power Respiration 4 Uses the electron transport chain to convert reducing power to proton motive force Yields ATP by oxidative phosphorylation 2 NADH Oxidative phosphorylation Substrate-level phosphorylation Pyruvate Pyruvate AcetylCoA AcetylCoA 2 NADH ~~ 6 ATP 6 NADH ~~ 18 ATP ~~ 4 ATP Oxidative phosphorylation x2 CO2 Oxidative phosphorylation 2 FADH2 CO2 TCA cycle Incorporates an acetyl group and releases CO2 (TCA cycles twice) ~~ 6 ATP ~~ 2 ATP Oxidative phosphorylation Substrate-level phosphorylation ~~ 2 ATP The Electron Transport Chain—Generating Proton Motive Force § Calculating theoretical maximum yields • In prokaryotes: • • • • Glycolysis: 2 NADHà 6 ATP Transition step: 2 NADH à 6 ATP TCA Cycle: 6 NADH à 18 ATP; 2 FADH2 à 4 ATP Total maximum oxidative phosphorylation yield = 34 ATP • Slightly less in eukaryotic cells • NADH from glycolysis in cytoplasm transported across mitochondrial membrane to enter electron transport chain – Requires ~1 ATP per NADH generated 13 The Electron Transport Chain—Generating Proton Motive Force § ATP Yield of Aerobic Respiration in Prokaryotes • Substrate-level phosphorylation: • 2 ATP (from glycolysis; net gain) • 2 ATP (from the TCA cycle) • 4 ATP (total) • Oxidative phosphorylation: • • • • 6 ATP (from reducing power gained in glycolysis) 6 ATP (from reducing power gained in transition step) 22 ATP (from reducing power gained in TCA cycle) 34 (total) • Total ATP gain (theoretical maximum) = 38 Overview of Catabolism § Fermentation • If cells cannot respire, will run out of carriers available to accept electrons • Glycolysis will stop • Fermentation uses pyruvate or derivative as terminal electron acceptor to regenerate NAD+ • Glycolysis can continue 6.5. Fermentation § Fermentation used when respiration not an option Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. • E. coli is facultative anaerobe • Aerobic respiration, anaerobic respiration, and fermentation GLUCOSE Pentose phosphate pathway Starts the oxidation of glucose 2 Yields Glycolysis Oxidizes glucose to pyruvate 1 P ~ P ~P + Reducing power ATP by substrate-level phosphorylation Yields Reducing power Biosynthesis Fermentation Reduces pyruvate or a derivative 5 Pyruvate Pyruvate 3a Acids, alcohols, and gases Transition step CO2 CO2 Yields Reducing power AcetylCoA AcetylCoA • Streptococcus pneumoniae lacks electron transport chain x CO2 2 CO2 3b TCA cycle Incorporates an acetyl group and releases CO2 (TCA cycles twice) Yields P ~ P ~ Reducing power P ATP by substrate-level phosphorylation + 4 • Fermentation only option P ~ P ~ P ATP by oxidative phosphorylation • ATP-generating reactions are only those of glycolysis • Additional steps consume excess reducing power – Regenerate NAD+ Respiration Uses the electron transport chain to convert reducing power to proton motive force Yields NADH + H+ NAD+ H 3C O O C C O– H 3C OH O C C O– H Lactate Pyruvate (a) Lactic acid fermentation CO2 H 3C O O C C O– Pyruvate NADH+ H+ O H 3C C H Acetaldehyde NAD+ OH H 3C C H H Ethanol (b) Ethanol fermentation 14 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. GLUCOSE Pentose phosphate pathway Starts the oxidation of glucose 2 Yields Glycolysis Oxidizes glucose to pyruvate 1 P ~ P ~ Reducing power + P ATP by substrate-level phosphorylation Yields Reducing power Biosynthesis Fermentation Reduces pyruvate or a derivative 5 Pyruvate Pyruvate Acids, alcohols, and gases 3a Transition step CO2 CO2 Yields Reducing power AcetylCoA AcetylCoA x CO2 2 CO2 3b TCA cycle Incorporates an acetyl group and releases CO2 (TCA cycles twice) Yields P ATP by substrate-level phosphorylation ~ P ~ Reducing power P + 4 Respiration Uses the electron transport chain to convert reducing power to proton motive force Yields P ~ P ~ P ATP by oxidative phosphorylation NADH + H 3C O O C C NAD+ H+ O– OH O C C H 3C O– H Lactate Pyruvate (a) Lactic acid fermentation CO2 O H 3C C + NADH O H+ NAD+ O O– C Pyruvate H 3C OH C H 3C H C H H Ethanol Acetaldehyde (b) Ethanol fermentation 6.5. Fermentation § Fermentation end products varied; helpful in identification, commercially useful • Ethanol • Butyric acid • Propionic acid • 2,3-Butanediol • Mixed acids Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Pyruvate Fermentation pathway Microorganisms End products Lactic acid Ethanol Butyric acid Propionic acid Mixed acids 2,3-Butanediol Streptococcus Lactobacillus Saccharomyces Clostridium Propionibacterium E. coli Enterobacter Lactic acid Ethanol CO2 Butyric acid Butanol Acetone Isopropanol CO2 H2 Propionic acid Acetic acid CO2 Acetic acid Lactic acid Succinic acid Ethanol CO2 H2 CO2 H2 (yogurt, dairy, pickle), b (wine, beer), (acetone): © Brian Moeskau/McGraw- Hill; (cheese): © Photodisc/McGraw-Hill; (Voges-Proskauer Test), (Methyl-Red Test): © The McGraw-Hill Companies, Inc./Auburn University Photographic Services 6.6. Catabolism of Organic Compounds Other than Glucose § Microbes can use variety of compounds • Excrete hydrolytic enzymes; transport subunits into cell • Degrade further to appropriate precursor metabolites • Polysaccharides and disaccharides • Amylases digest starch; cellulases digest cellulose • Disaccharides hydrolyzed by specific disaccharidases • Lipids • Fats hydrolyzed by lipases; glycerol converted to dihydroxyacetone phosphate, enters glycolysis • Fatty acids degraded by β-oxidation to enter TCA cycle • Proteins • Hydrolyzed by proteases; amino group deaminated • Carbon skeletons converted into precursor molecules 15 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Fig. 6.24 POLYSACCHARIDES Starch Cellulose amylases cellulases DISACCHARIDES Lactose Maltose Sucrose LIPIDS (fats) proteases Amino acids + monosaccharides (simple sugars) Pentose phosphate pathway PROTEINS lipases glycerol disaccharidases GLUCOSE deamination fatty acids NH3 Glycolysis Applies to both branches In glycolysis Pyruvate Pyruvate AcetylCoA AcetylCoA ß-oxidation removes 2-carbon units. x2 TCA cycle 6.7. Chemolithotrophs § Prokaryotes unique in ability to use reduced inorganic compounds as sources of energy • E.g., hydrogen sulfide (H2S), ammonia (NH3) • Produced by anaerobic respiration from inorganic molecules (sulfate, nitrate) serving as terminal electron acceptors • Important example of nutrient cycling • Four general groups 6.10. Anabolic Pathways—Synthesizing Subunits from Precursor Molecules § Prokaryotes remarkably similar in biosynthesis • Synthesize subunits using central metabolic pathways • If enzymes lacking, end product must be supplied • Fastidious bacteria require many growth factors • Lipid synthesis requires fatty acids, glycerol • Fatty acids: 2-carbon units added to acetyl group from acetyl-CoA • Glycerol: dihydroxyacetone phosphate from glycolysis • Nucleotide synthesis • • • • DNA, RNA initially synthesized as ribonucleotides Purines: atoms added to ribose 5-phosphate to form ring Pyrimidines: ring made, then attached to ribose 5-phosphate Can be converted to other nucleobases of same type 16