* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Regulation of Gene Expression
Genome evolution wikipedia , lookup
Gene desert wikipedia , lookup
Community fingerprinting wikipedia , lookup
X-inactivation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Molecular evolution wikipedia , lookup
List of types of proteins wikipedia , lookup
Non-coding DNA wikipedia , lookup
Messenger RNA wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
RNA interference wikipedia , lookup
Gene regulatory network wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Polyadenylation wikipedia , lookup
Deoxyribozyme wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Epitranscriptome wikipedia , lookup
RNA silencing wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Gene expression wikipedia , lookup
Non-coding RNA wikipedia , lookup
Transcriptional regulation wikipedia , lookup
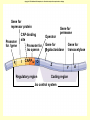
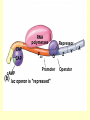
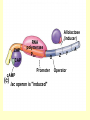
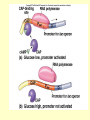
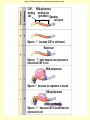
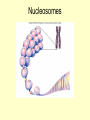
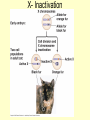
Control of Transcription DNA has “on” and “off” switches Activator –protein that binds near gene’s promoter region - allows RNA polymerase to transcribe (allows it to fit) Repressor – protein that binds to DNA and prevents RNA polymerase from binding -coded for by “regulator” gene The Operon Model Operon – region of DNA with group of genes for proteins with related functions (see diagram) -All of the genes in the operon are controlled by activity at the promoter & the operator Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Gene for repressor protein Promoter for I gene PI CAP-binding site Promoter for lac operon I CAP Plac O Regulatory region Operator Gene for permease Gene for b-galactosidase Z Gene for transacetylase Y Coding region lac control system A The “Lac” Operon -in E. coli bacteria -genes for enzymes to break down lactose sugar 1. Regulator gene codes for Lac repressor protein 2. Repressor binds with operator region If no lactose present: 3. RNA polymerase can not bind to promoter, no enzyme made and operon is “off” If lactose is present: 3. Lactose binds with repressor, repressor no longer binds to operator 4. RNA polymerase transcribes the structural genes 5. Translation occurs, enzymes are made 6. Lactose is metabolized *Conservation of resources – enzymes only made when needed* Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. RNA polymerase CAP I cAMP CAP Plac O Promoter lac operon is "repressed" Repressor Y Z Operator A Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. CAP I RNA polymerase Plac CAP Allolactose (inducer) O Promoter cAMP lac operon is "induced" Z Y Operator A Glucose Lactose Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. + + RNA-polymrase CAPbinding binding site (promoter) Operator site lacZ gene Operon ? because CAP is not bound Repressor + – Operon ? both because lac repressor is bound and CAP is not RNA polymerase – – CAP Operon ? because lac repressor is bound RNA polymerase – + CAP mRNA synthesis Operon ? because CAP is bound and lac repressor is not RNA in Gene regulation “Small RNA’s” Process of RNA interference (RNAi) or “RNA silencing” -double stranded RNA is cut into small pieces by enzyme called dicer -pieces then unravel into single strands 1. miRNA – micro RNA -binds to a specific RNA thus blocking it from being translated -reversible 2. siRNA – small interfering RNA -binds to specific mRNA, destroys it Purposes: (evolutionary origins) 1. Inactivate viral DNA transcription 2. Inactivate transposons – renegade “jumping genes” 3. Regulation Nucleosomes X- Inactivation