* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download ppt - Sol Genomics Network
Gene therapy of the human retina wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Non-coding DNA wikipedia , lookup
Ridge (biology) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy wikipedia , lookup
Oncogenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genetic engineering wikipedia , lookup
Transposable element wikipedia , lookup
Copy-number variation wikipedia , lookup
Metagenomics wikipedia , lookup
Gene nomenclature wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene desert wikipedia , lookup
Human genome wikipedia , lookup
Public health genomics wikipedia , lookup
Genomic library wikipedia , lookup
Pathogenomics wikipedia , lookup
Microevolution wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genome (book) wikipedia , lookup
Minimal genome wikipedia , lookup
Helitron (biology) wikipedia , lookup
Human Genome Project wikipedia , lookup
Designer baby wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
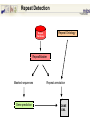
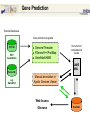
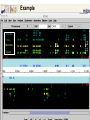
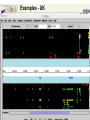
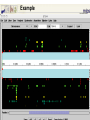
A Bioinformatic Framework to Unravel the Secrets of the Tomato Genome 15 January 2006, PAG XIV SanDiego Rémy Bruggmann, MIPS/IBI, GSF Outline Introduction Data management Annotation Training/Test gene set Summary MIPS´ look at the Green Side of Life – genome projects and database activities – Arabidopsis thaliana Arabidopsis lyrata * Capsella rubella * Maize Rice Medicago Lotus Solanum lycopersicum MIPS´ look at the Green Side of Life – genome projects and database activities – Need to streamline and unify databases as well as analytical schemas and operation routines Strong synergism and very robust Risk to loose flexibility and „custom tailor“ attractiveness Awareness that not every genome and every community „is just the same“ From Center Centric Strategies to distributed Approaches Typically, genome projects undergo particular phases: Sequenced BACs are annotated Gene models are published to the community Potentially generates competition rather than collaboration among groups From Center Centric Strategies to distributed Approaches Consequences can be: underlying analytical procedures are not always tested, trained and evaluated Between groups more or less pronounced differences exist --> differing, contradicting and confliciting data Aim of all groups: „information enriched high quality genome backbone to address genome scale biological questions“ From Center Centric Strategies to distributed Approaches An example ... International Medicago Genome Annotation Group Consists of groups participating either in the International or the European Medicago Genome Initiative annotation/ bioinformatics programs Agreement on common annotation standards, data exchange formats and naming conventions Aims to produce and provide unified high-quality Medicago data set From Center Centric Strategies to distributed Approaches Advantages of sharing efforts in genome annotation within a common annotation pipeline From Center Centric Strategies to distributed Approaches prevents from: (i) duplicating efforts (ii) conflicts resulted from different annotation “standards” ensures high-quality annotation standards ensures common (gene) naming common dataset Integrates and profits from knowledge and expertise of the individual groups Data management All data should be organized in a genome database Wishlist for a modern genome db Complete Comprehensive Up-to-date Integrated User interface Application interface State-of-the-art automatic analysis Adaptable Cross-genome comparison …low cost, low manpower... PlantsDB Philosophy Plants Genome Resource: provides and integrates sequence data from European plant sequencing consortia along with publically available data from the international initiative Plants DB communicates bioinformatic analysis data (visualization, genetic elements, structural data, ontologies, domains...; BLAST, browse and search,…comparative analysis) Integration: provides a distributed network to integrate and retrieve data from heterogenous resources using BioMOBY (connection to other plant DBs, PlaNET) Preliminary Annotation Pipeline Towards a preliminary annotation Repeat Detection Repeat Ontology Repeat Database RepeatMasker Masked sequences Gene prediction Repeat annotation GAME XML Gene Prediction External Databases Gene prediction programs EST DB EST Assemblies ► GenomeThreader ► FGenesH++/ProtMap Document of computational results ► GeneMarkHMM GAME XML Protein DB e.g. SwissProt Manual annotation in Apollo Genome Viewer Web Access Gbrowse PlantsDB First Results Repeat Masker [%] 5.8 MB analysed (48 BACs) 25 ~ 6.7 % repetitive elements (<0.2% - 23% per bac) 20 ~ 1 min/100 kb 15 Repeat content 10 whole genome (euchromatic part): 5 ~ 2 days 0 BACs State: December 2005 Preliminary Results Comparison of different gene finders ab initio predictions EST/TC GeneMark FGeneSH EST/TC ab initio predictions ab initio predictions FGeneSH++ and GeneMarkHMM often generate incomplete or wrong gene models at the moment There are no matrices available that are trained for tomato Tomato matrices will increase prediction quality dramatically Collection of annotated high quality genes for a training/test set for EuGene, FGeneSH, GeneMarkHMM, ... Training/Test Gene Set How can we get a training/test set? Map available tomato cDNA/ESTs to the BACs (use only high confident matches) Link experimental data to the genemodels Use this gene set for ab initio gene finder training GenomeThreader GenomeThreader used for EST/cDNA-Mapping: similarity-based approach: EST/Proteins used to predict gene structure via optimal spliced alignments Offers many options (full user control) incremental updates (avoids a lot of duplicated computations) Improved GeneSeqer GenomeThreader - calculations DB Entries Size [MB] Calc time/100kb [s] Whole Genome Tomato 32401 27 27 s MicroTom 26363 21 22 s Potato 38239 34 23 s Tobacco 28661 20 39 s Arabidopsis cDNAs 31939 45 10 s 0.3 days 404822 311 170 s 4.3 days 15639 21 8s 0.2 days Uni_trembl Plants 185564 74 38 s 1.0 day Uniprot_swissprot 181571 82 8s 0.2 days Nonred 1675230 662 437 s 11.1 days Total 2834224 1433 14 min 22 days Dicots rice cds ~ 2.8 days (single CPU, euchromatic part) Example Tobacco Potato Microtom Tomato Examples - UK Example Number of high quality genes # genes Number of genes: 164 (covered completely by cDNA/ESTs) 10 8 6 ~3.4 genes/BAC (range: 0 - 9 genes/BAC) 4 2 These genes can be used to train gene finders (Only very good alignments considered) 0 BAC Gene Finder Which program can be trained for tomato? One possibility is EuGene (VIB Gent) - performed well e.g. for Arabidopsis and Medicago - available as soon as test/training gene set is large enough EuGene - overview Plugins Statistical contents DNA Markov AA Markov Splice sites NetGene2 GeneSplicer SpliceMachine SplicePredictor Start sites SpliceMachine NetStart ATRPred Similarities EST similarities Protein similarities FL cDNA Repeats Exon conservation Plugin training Optimize plugin Test combination Needs Needs Needs one one one dataset dataset new dataset TRAINING OPTIM TEST EuGene First round training: - 500 high quality tomato genes - statistical models on codon usage and splice sites of Arabidopsis will be used Second round training: - 2000 high quality tomato genes - Build a tomato-only version of EuGene Approx. 150 BACs needed for first round training Current state of sequenced BACs Total number of BACs: - unfinished: 71 - finished: 87 - available: 52 Summary ab initio gene finders are not yet calibrated to tomato Need of a test/training gene set to calibrate the gene finders We need another 100 BACs to get enough genes for a first round training of EuGene GenomeThreader produces only good alignments with ESTs from SOL-species (Tomato, Potato, Tobacco) More repeats will be detected (will be included in RepeatMasker Library) Acknowledgments Automated annotation MIPS Heidrun Gundlach Georg Haberer Manuel Spannagl Klaus F.X. Mayer Manual Annotation/Curation/Web-site (Chromosome 4) Imperial College Daniel Buchan James Abbot Sarah Butcher Gerard Bishop Sequencing & Assembly (Chromosome 4) Sanger Institute Christine Nicholson Sean Humphray MPIZ Köln Heiko Schoof EuGene VIB Gent Stephane Rombauts GenomeThreader University of Hamburg Gordon Gremme Stefan Kurtz Volker Brendel A Bioinformatic Framework to Unravel the Secrets of the Tomato Genome 15 January 2006, PAG XIV SanDiego Rémy Bruggmann, MIPS/IBI, GSF