* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download cDNA Micoroarray Data Analysis
Promoter (genetics) wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Gene regulatory network wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Gene expression wikipedia , lookup
Signal transduction wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein moonlighting wikipedia , lookup
Expression vector wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Biochemical cascade wikipedia , lookup
Interactome wikipedia , lookup
List of types of proteins wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Western blot wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
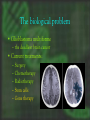
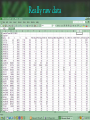
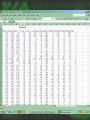
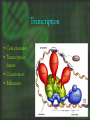
cDNA Microarray analysis of an invasive brain tumor OR More answers than you can handle Dominique B Hoelzinger Overview I. II. III. IV. Introduction Generating data Analyzing data Interpreting data The biological problem • Glioblastoma multiforme – the deadliest brain cancer • Current treatments: – – – – – Surgery Chemotherapy Radiotherapy Stem cells Gene therapy SPREAD OF GLIOBLASTOMA MULTIFORME 1) corpus callosum 2) Fornix 3) Optic radiation 4) Association pathways 5) Anterior commissure Glioma motility • What make these cells move? • What switches them from dividing to motile? The ones that got away • Highly invasive – Surgeon can’t reach them – Chemotherapy and radiotherapy can’t reach them – They are not dividing core rim core rim Laser Capture Microdissection 1) Prepare Follow routine protocols for preparing a tissue on a plain, uncovered microscope slide 2) Locate Visualize the sample through the video monitor or the microscope. Position the CapSure™ film carrier over the cell(s) of interest 3) Capture Press the button to pulse the low power infrared laser. The desired cell(s) adhere to the CapSure ™ film carrier. 4) Microdissect Lift the CapSure ™ film carrier, with the desired cell(s) to the film surface. The surrounding tissue remains intact. Place the CapSure ™ film carrier directly onto a standard microcentrifuge tube (Eppendorf) containing the extraction buffer. The cell contents, DNA, RNA or are ready for subsequent molecular analysis. 5) Analyze Microdissection of single cells • Identify invading glioma cells on cryostat sections • Using 20x magnification, laser-capture tumor cells • Retrieve captured cells on LCM Cap • Verify cell capture by inspection of Cap 10mm About RNA Overview I. II. III. IV. Introduction Generating data Analyzing data Interpreting data Robotic Array Assembly cDNA microarray technology http://research.nhgri.nih.gov/microarray/image_analysis.html Really raw data Overview I. II. III. IV. Introduction Generating data Analyzing data Interpreting data GeneSpring • Normalizes the calculated data • Selects genes more than twofold over or under the ratio of 1 (equally expressed in both populations) • Custer analysis • Principal Components Analysis Genes down-regulated in migrating cells • C/R Name Description • Extracellular • • • • • • • • • • • 33 12 11 11 10 7 5 4 4 3 3 • Vascular Involvement/Angiogenesis • • • • • • • • 43 FCGR3A Fc fragment of IgG, low affinity IIIa, 42 PTGER4 prostaglandin E receptor 4 (subtype 17 HLA-DRA major histocompatibility complex, class II, 6 CD163 CD 163 antigen 5 VEGF vascular endothelial growth factor 5 VCAM1 vascular cell adhesion molecule 1 4 LMO2LIM domain only 2 (rhombotin-like1) 4 CD68 CD68 antigen Signal Transduction • • • • • • • 6 8 4 3 3 3 3 IGFBP5 insulin-like growth factor binding protein 5 IGFBP2 insulin-like growth factor binding protein 2 DEPP decidual protein induced by progesterone ABCC3 ATP-binding cassette, C (CFTR/MRP) 3 TNC tenascin C (hexabrachion) SRPX sushi-repeat-containing protein, X chrom SFRP4secreted frizzled-related protein 4 SERPINB2 serine (or cystein) proteinase inhibitor, 2 (P SERPINH2 serine (or cystein) proteinase inhibit MUC1 mucin 1 EGFR-RS Likely ortholog of mouse EGF IQGAP IQ motiv containing GTPase activating RDC1 G protein-coupled receptor RGS16 Regulator of G-protein signaling 16 NFKBIA NFKB inhibitor, alpha PLD2 phospholipase D 2 TK2 thymidine kinase 2, mitochondrial ABL1 abelson murine leukemia viral oncogene homolog 1 Cytoskeleton 12 VIM vimentin 7 PLEK plekstrin 5 MSN moesin 4 CAPG Capping protein (actin filament), gelsolin-like 3 KANK kidney ankyrin repeat-containing protein Apoptosis 4 CASP4 caspase 4 4 PIG3 p53 induced gene 3 Transcription 14 FP36L1 zinc finger protein 36, C3H type-like 1 (ERF-1) 7 ID4 inhibitor of DNA binding 4, dominant neg helix-loop-helix protein 3 BTF3 basic transcription factor 3 6 EYA2 eyes absent (Drosophila) homolog 2 4 EGR1 Early growth response 1 4 JUNB Jun B proto-oncogene 4 CEBPB CCAAT/enhancer binding protein (C/EBP), beta 3 NFKBIA nuclear factor kappa-B inhibitor alpha 3 FOXM1 forkhead box 1M Proliferation 3 CKS2 CDC28 protein kinase regulatory subunit 2 3 CDC20 cell division cycle 20 Unknown function 5 H47315 EST 7 MT1L metallothionein 1L 6 CLIC1 chloride intracellular channel 1 6 MT2A metallothionein 2A 4 HNRPH1 heterogeneous nuclear ribonucleoprotein H1 4 R68464 EST 4 APOE apolipoprotein E 3 KIAA0630 KIAA0630 protein 3 MSI2 Musashi homolog 2 Overview I. II. III. IV. Introduction Generating data Analyzing data Interpreting data BioHavasu project Unusual Suspects: Cataloging Cancer Related Proteins, Genes using Biomedical Literature • • • • Pathway involvement (activity of protein): Determine the cellular pathway(s) during which the protein is involved : apoptosis, proliferation, or migration Interaction (protein/protein , protein/nucleic acids or protein /fatty acids): Determine protein binding. Swissprot, Entrez protein or Expasy Disease (protein/disease, protein/tissue type): Determine the types of cancer that the protein is related to. Protein Action (protein/function): Determine the diverse activation and inhibition relationships between proteins as well as sub-cellular localization. Understanding relationships Laminin a SERPIN B2 5 DKK3 Elastin SFRP4 HGF/SF VCAM IGFBP2 TNC FGF 9 VEGF KLK6 Ephrin- B2 SERPINH2 Collagen IX IGFBP5 SPOCK tenascinC LPA ENPP2 PTPRN2 EFNB3 OPCML 2 EGFR-RS EGFR a Eph B6 RGS16 DTR GPCRs b g GRIA1 integrins paxillin G proteins FAK STX11 RGS7 Ras IQGAP Actin Guanine exchange factors PKCB PKC Rac Cdc42 ARHGAP8 Rho Rho ROCK Transcription factors Pak ZFP36L1, ID4, BTF3, EYA2, EGR1, JUNB WASP DAP3,BCL2L2 MLCK AP3M2 Up-regulated during invasion Apoptosis MLC phosphatase LIM kinase TRABID, MEF2C, ETS2, BACH2 Down-regulated during invasion CAPG Nucleation of actin at membrane Cofilin Actin depolymerization CASP4, PIG3 myosin Retrograde flow of actin filaments profilin Actin polymerization stress fibers Sub-cellular localization Proposed Ontology-Directed Extraction Methodology • Model Medical Terminology: Identify existing medical ontologies such as UMLS for modeling the domain knowledge. • Text Classifier Module: Build a classifier for identifying “interesting” sentences in MEDLINE abstracts. • Natural Language Processing: Identify pre-processing steps for structuring free-text. Such steps involve part of speech tagging, noun and verb phrase chunking and shallow parsing. • Relationship Extractor Module: Build an extractor system using machine-learning techniques, such as ILP, for learning rules that combine the medical ontologies with learned patterns on sentences to extract relationships among proteins. • Usability, Performance and Scalability: Determine if the system is usable by biologists, if it can be easily trained to extract new types of relationships and its recall and precision is at acceptable levels. So that I don’t have to spend hours finding diagrams myself…. Mef 2C LPA GCR G proteins HB-EGF Promoter Analysis • Find the promoter region – Genome browser • Find transcription binding site – TESS – Genomatix – Biobase, etc • Align several promoters to find common patterns The ones that got away • Highly invasive – Surgeon can’t reach them – Chemotherapy and radiotherapy can’t reach them – They are not dividing core rim core rim Genetics again! Transcription • Core promoter • Transcription factors • Co-activators • Enhancers Transcription factors Consensus binding sites • Position weighted matrices – Define variation in promoter consensus sequences The sequenced human genome Finding the Promoter Genome Browser Human Genome Browser Gateway TESS TESS Job W0793006061 : Tabulated Results Promoter structure 1 2 3 4 Promoter Alinement Genomatix The next step, biological significance • Proof of transcriptional regulation = proof of protein – Cellular specificity – Subcellular localization – Activity TissueInformatics Tissue micro-array Conclusion • cDNA microarray technology has opened a flood gate of information • Biologists need HELP • Expedite the interpretation of data. • ideas wanted