* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Brooker Chapter 14
Transcription factor wikipedia , lookup
Magnesium transporter wikipedia , lookup
Non-coding RNA wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Molecular evolution wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Western blot wikipedia , lookup
Protein adsorption wikipedia , lookup
Gene expression profiling wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Protein moonlighting wikipedia , lookup
Epitranscriptome wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Proteolysis wikipedia , lookup
Point mutation wikipedia , lookup
Expression vector wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene regulatory network wikipedia , lookup
Gene expression wikipedia , lookup
Transcriptional regulation wikipedia , lookup
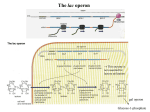
PowerPoint Presentation Materials to accompany Genetics: Analysis and Principles Robert J. Brooker CHAPTER 14 GENE REGULATION IN BACTERIA AND BACTERIOPHAGES Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display INTRODUCTION The term gene regulation means that the level of gene expression can vary under different conditions Genes that are unregulated are termed constitutive They have essentially constant levels of expression Frequently, constitutive genes encode proteins that are necessary for the survival of the organism The benefit of regulating genes is that encoded proteins will be produced only when required Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-2 INTRODUCTION Gene regulation is important for cellular processes such as 1. Metabolism 2. Response to environmental stress 3. Cell division Regulation can occur at any of the points on the pathway to gene expression Refer to Figure 14.1 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-3 Figure 14.1 14-4 14.1 TRANSCRIPTIONAL REGULATION The most common way to regulate gene expression in bacteria is at the transcriptional level The rate of RNA synthesis can be increased or decreased Transcriptional regulation involves the actions of two main types of regulatory proteins Repressors Bind to DNA and inhibit transcription Activators Bind to DNA and increase transcription Negative control refers to transcriptional regulation by repressor proteins Positive control to regulation by activator proteins Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-5 Small effector molecules affect transcription regulation However, these bind to regulatory proteins and not to DNA directly In some cases, the presence of a small effector molecule may increase transcription These molecules are termed inducers They function in two ways Bind activators and cause them to bind to DNA Bind repressors and prevent them from binding to DNA Genes that are regulated in this manner are termed inducible In other cases, the presence of a small effector molecule may inhibit transcription Corepressors bind to repressors and cause them to bind to DNA Inhibitors bind to activators and prevent them from binding to DNA Genes that are regulated in this manner are termed repressible Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-6 Regulatory proteins have two binding sites Figure 14.2 One for a small effector molecule The other for DNA 14-7 Figure 14.2 14-8 The Phenomenon of Enzyme Adaptation At the turn of the 20th century, scientists made the following observation A particular enzyme appears in the cell only after the cell has been exposed to the enzyme’s substrate This observation became known as enzyme adaptation François Jacob and Jacques Monod at the Pasteur Institute in Paris were interested in this phenomenon They focused their attention on lactose metabolism in E. coli to investigate this problem Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-9 The lac Operon An operon is a regulatory unit consisting of a few structural genes under the control of one promoter It encodes polycistronic mRNA that contains the coding sequence for two or more structural genes This allows a bacterium to coordinately regulate a group of genes that encode proteins with a common function An operon contains several different regions Promoter; terminator; structural genes; operator Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-10 Figure 14.3a shows the organization and transcriptional regulation of the lac operon genes There are two distinct transcriptional units 1. The actual lac operon a. DNA elements Promoter Binds RNA polymerase Operator Binds the lac repressor protein CAP site Binds the Catabolite Activator Protein (CAP) b. Structural genes lacZ Encodes b-galactosidase Enzymatically cleaves lactose and lactose analogues Also converts lactose into allolactose (an isomer) lacY Encodes lactose permease Membrane protein required for transport of lactose and analogues lacA Encodes transacetylase Covalently modifies lactose and analogues Its functional necessity remains unclear Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-11 Figure 14.3a shows the organization and transcriptional regulation of the lac operon genes There are two distinct transcriptional units 2. The lacI gene Not considered part of the lac operon Has its own promoter, the i promoter Constitutively expressed at fairly low levels Encodes the lac repressor The lac repressor protein functions as a tetramer Only a small amount of protein is needed to repress the lac operon There is usually ten tetramer proteins per cell Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-12 Figure 14.3 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-13 The lac Operon Is Regulated By a Repressor Protein The lac operon can be transcriptionally regulated 1. By a repressor protein 2. By an activator protein The first method is an inducible, negative control mechanism It involves the lac repressor protein The inducer is allolactose It binds to the lac repressor and inactivates it Refer to Figure 14.4 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-14 RNA pol cannot access the promoter Constitutive expression The lac operon is now repressed Therefore no allolactose Figure 14.4 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-15 Translation The lac operon is now induced The conformation of the repressor is now altered Repressor can no longer bind to operator Some gets converted to allolactose Figure 14.4 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-16 Repressor does not completely inhibit transcription So very small amounts of the enzymes are made Figure 14.5 The cycle of lac operon induction and repression Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-17 Experiment 14A: The lacI Gene Encodes a Repressor Protein In the 1950s, Jacob and Monod, and their colleague Arthur Pardee, had identified a few rare mutant strains of bacteria with abnormal lactose adaptation One type of mutant involved a defect in the lacI gene It was designated lacI– It resulted in the constitutive expression of the lac operon even in the absence of lactose The lacI– mutations mapped very close to the lac operon Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-18 Jacob, Monod and Pardee proposed two different functions for the lacI gene Figure 14.6 Jacob, Monod and Pardee applied a genetic approach to distinguish between the two hypotheses Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-19 They used bacterial conjugation methods to introduce different portions of the lac operon into different strains They identified F’ factors (plasmids) that carried portions of the lac operon For example: Consider an F’ factor that carries the lacI gene Bacteria that receive this will have two copies of the lacI gene One on the chromosome and the other on the F’ factor These are called merozygotes, or partial diploids Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-20 Merozygotes were instrumental in allowing Jacob et al to elucidate the function of the lacI gene There are two key points 1. The two lacI genes in a merozygote may be different alleles lacI– on the chromosome lacI+ on the F’ factor 2. Genes on the F’ factor are not physically connected to those on the bacterial chromosome If hypothesis 1 is correct The repressor protein produced from the F’ factor can diffuse and regulate the lac operon on the bacterial chromosome If hypothesis 2 is correct The binding site on the F’ factor cannot affect the lac operon on the bacterial chromosome, because they are not physically adjacent Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-21 The Hypothesis The lacI gene either/or 1. Encodes a regulatory protein (the lac repressor) that can diffuse throughout the cell 2. Acts as a binding site for a repressor protein Thus it can only act on a physically connected lac operon Testing the Hypothesis Refer to Figure 14.7 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-22 Figure 14.7 14-23 Figure 14.7 14-24 Figure 14.7 14-25 The Data Strain Addition of lactose Amount of b-galactosidase (percentage of parent strain) Mutant No 100% Mutant Yes 100% Merozygote No <1% Merozygote Yes 220% Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-26 Interpreting the Data Strain Addition of lactose Expected result because of constitutive expression in the lacI– strain Amount of b-galactosidase (percentage of parent strain) Mutant No 100% Mutant Yes 100% Merozygote No <1% Merozygote Yes 220% In the presence of lactose, both lac operons are induced, yielding a higher level of enzyme activity In the absence of lactose, both lac operons are repressed This result is consistent with hypothesis 1 The lacI gene codes for a repressor protein that can diffuse throughout the cell and bind to any lac operon Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-27 Interpreting the Data The interaction between regulatory proteins and DNA sequences have led to two definitions 1. Trans-effect Genetic regulation that can occur even though DNA segments are not physically adjacent Mediated by genes that encode regulatory proteins Example: The action of the lac repressor on the lac operon 2. Cis-effect or cis-acting element A DNA sequence that must be adjacent to the gene(s) it regulates Mediated by sequences that bind regulatory proteins Example: The lac operator Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-28 Interpreting the Data Table 14.1 summarizes the effects of lacI gene mutations versus lacO (operator) in merozygotes Overall A mutation in a trans-acting factor is complemented by the introduction of a second gene with a normal function However, a mutation in a cis-acting element is not! Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-29 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-30 The lac Operon Is Also Regulated By an Activator Protein The lac operon can be transcriptionally regulated in a second way, known as catabolite repression When exposed to both lactose and glucose E. coli uses glucose first, and catabolite repression prevents the use of lactose When glucose is depleted, catabolite repression is alleviated, and the lac operon is expressed The sequential use of two sugars by a bacterium is termed diauxic growth Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-31 The lac Operon Is Also Regulated By an Activator Protein The small effector molecule in catabolite repression is not glucose This form of genetic regulation involves a small molecule, cyclic AMP (cAMP) It is produced from ATP via the enzyme adenylyl cyclase cAMP binds an activator protein known as the Catabolite Activator Protein (CAP) Also termed the cyclic AMP receptor protein (CRP) Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-32 The lac Operon Is Also Regulated By an Activator Protein The cAMP-CAP complex is an example of genetic regulation that is inducible and under positive control The cAMP-CAP complex binds to the CAP site near the lac promoter and increases transcription In the presence of glucose, the enzyme adenylyl cyclase is inhibited This decreases the levels of cAMP in the cell Therefore, cAMP is no longer available to bind CAP Transcription rate decreases Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-33 (b) Lactose but no cAMP Figure 14.8 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-34 Figure 14.8 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-35 The lac Operon Has Three Operator Sites for the lac Repressor Detailed genetic and crystallographic studies have shown that the binding of the lac repressor is more complex than originally thought In all, three operator sites have been discovered O1 Next to the promoter O2 Downstream in the lacZ coding region O3 Slightly upstream of the CAP site Refer to Figure 14.9 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-36 Repression is 1,300 fold Therefore, transcription is 1/1,300 the level when lactose is present No repression ie: Constitutive expression Figure 14.9 The identification of three lac operator sites 14-37 The results of Figure 14.9 supported the hypothesis that the lac repressor must bind to two of the three operators to cause repression It can bind to O1 and O2 , or to O1 and O3 If either O2 or O3 is missing maximal repression is not achieved Binding of the lac repressor to two operator sites requires that the DNA form a loop A loop in the DNA brings the operator sites closer together But not O2 and O3 This facilitates the binding of the repressor protein Refer to Figure 4.14 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-38 Each repressor dimer binds to one operator site Each repressor dimer binds to one operator site Figure 14.10 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-39 The ara Operon Another operon in E. coli that is involved in sugar metabolism is the ara (arabinose) operon It contains Three structural genes involved in arabinose metabolism These are designated araB, araA and araD A single promoter, PBAD A CAP site, which binds the catabolite activator protein Refer to Figure 14.11 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-40 The araC gene is adjacent to the ara operon It has its own promoter, PC It encodes a regulatory protein, AraC AraC can bind to three different operator sites Designated araI, araO1 and araO2 The AraC protein can act as either a negative or positive regulator of transcription Depending on whether or not arabinose is present Figure 14.11 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-41 AraC protein binds to all three operators AraC dimer bound to araO1 inhibits transcription of the araC gene This keeps AraC protein levels fairly low AraC monomers bound to araO2 and araI repress the ara operon They bind to each other (via looped DNA), and block RNA pol access to PBAD araO1 Figure 14.12 14-42 Arabinose binds to the AraC proteins The interaction betweem the AraC proteins at the araO2 and araI is broken This breaks the DNA loop Another, AraC protein binds to araI This AraC dimer at araI activates transcription CAP-cAMP activation occurs when glucose levels are low Figure 14.12 14-43 The trp Operon The trp operon (pronounced “trip”) is involved in the biosynthesis of the amino acid tryptophan The genes trpE, trpD, trpC, trpB and trpA encode enzymes involved in tryptophan biosynthesis The genes trpR and trpL are involved in regulation trpR Encodes the trp repressor protein Functions in repression trpL Encodes a short peptide called the Leader peptide Functions in attenuation Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-44 RNA pol can bind to the promoter Cannot bind to the operator site Figure 14.13 Organization of the trp operon and regulation via the trp repressor protein 14-45 Figure 14.13 Organization of the trp operon and regulation via the trp repressor protein 14-46 Another mechanism of regulation Figure 14.13 Organization of the trp operon and regulation via the trp repressor protein 14-47 Attenuation occurs in bacteria because of the coupling of transcription and translation During attenuation, transcription actually begins but it is terminated before the entire mRNA is made A segment of DNA, termed the attenuator, is important in facilitating this termination In the case of the trp operon, transcription terminates shortly past the trpL region (Figure 14.13c) Thus attenuation inhibits the further production of tryptophan The segment of trp operon immediately downstream from the operator site plays a critical role in attenuation The first gene in the trp operon is trpL It encodes a short peptide termed the Leader peptide Refer to Figure 14.14 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-48 Region 2 is complementary to regions 1 and 3 Region 3 is complementary to regions 2 and 4 Therefore several stem-loops structures are possible The 3-4 stem loop is followed by a sequence of Uracils It acts as an intrinsic (r-independent) terminator These two codons provide a way to sense if there is sufficient tryptophan for translation Figure 14.14 Sequence of the trpL mRNA produced during attenuation 14-49 Therefore, the formation of the 3-4 stem-loop causes RNA pol to terminate transcription at the end of the trpL gene Conditions that favor the formation of the 3-4 stem-loop rely on the translation of the trpL mRNA There are three possible scenarios 1. No translation 2. Low levels of tryptophan 3. High levels of tryptophan Refer to Figure 14.15 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-50 Transcription terminates just past the trpL gene Most stable form of mRNA occurs Therefore no coupling of transcription and translation Figure 14.15 Possible stem-loop structures formed from trpL mRNA under different conditions of translation 14-51 Region 1 is blocked 3-4 stem-loop does not form RNA pol transcribes rest of operon Insufficient amounts of tRNAtrp Figure 14.15 Possible stem-loop structures formed from trpL mRNA under different conditions of translation 14-52 Sufficient amounts of tRNAtrp 3-4 stem-loop forms Translation of the trpL mRNA progresses until stop codon RNA polymerase pauses Transcription terminates Region 2 cannot base pair with any other region Figure 14.15 Possible stem-loop structures formed from trpL mRNA under different conditions of translation 14-53 Inducible vs Repressible Regulation The study of many operons revealed a general trend concerning inducible versus repressible regulation Operons involved in catabolism (ie. breakdown of a substance) are typically inducible The substance to be broken down (or a related compound) acts as the inducer Operons involved in anabolism (ie. biosynthesis of a substance) are typically repressible The inhibitor or corepressor is the small molecule that is the product of the operon Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-54 14.2 TRANSLATIONAL AND POSTTRANSLATIONAL REGULATION Genetic regulation in bacteria is exercised predominantly at the level of transcription However, there are many examples of regulation that occur at a later stage in gene expression For example, regulation of gene expression can be Translational Posttranslational Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-55 Translational Regulation For some bacterial genes, the translation of mRNA is regulated by the binding of proteins A translational regulatory protein recognizes sequences within the mRNA In most cases, these proteins act to inhibit translation These are known as translational repressors Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-56 Translational Regulation Translational repressors inhibit translation in one of two ways 1. Binding next to the Shine-Dalgarno sequence and/or the start codon 2. Binding outside the Shine-Dalgarno/start codon region This will sterically hinder the ribosome from initiating translation They stabilize an mRNA secondary structure that prevents initiation Translational repression is also found in eukaryotes Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-57 Translational Regulation A second way to regulate translation is via the synthesis of antisense RNA An RNA strand that is complementary to mRNA Consider, for example, the trait of osmoregulation The ability to control the amount of water inside the cell The protein ompF in E. coli is important in osmoregulation This outer membrane protein is encoded by the ompF gene OmpF protein is preferentially produced at low osmolarity At high osmolarity its synthesis is decreased Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-58 The expression of another gene, termed micF, is responsible for inhibiting the ompF gene at high osmolarity micF RNA does not code for a protein It is, however, complementary to ompF mRNA It is thus termed antisense RNA Figure 14.16 Its translation is now blocked Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-59 Posttranslational Regulation A common mechanism to regulate the activity of metabolic enzymes is feedback inhibition The final product in a pathway often can inhibit the an enzyme that acts early in the pathway Refer to Figure 14.17 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-60 Enzyme 1 is an allosteric enzyme, with two different binding sites Catalytic site binds substrate Regulatory site binds final product of the pathway If the concentration of product 3 becomes high It will bind to enzyme 1 Thereby inhibiting its ability to convert substrate 1 into product 1 Figure 14.17 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-61 Posttranslational Regulation A second strategy to control the function of proteins is by the covalent modification of their structure Some modifications are irreversible Proteolytic processing Attachment of prosthetic groups, sugars, or lipids Other modifications are reversible and transiently affect protein function Phosphorylation (–PO4) Acetylation (–COCH3) Methylation (–CH3) Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-62 14.3 GENE REGULATION IN THE BACTERIOPHAGE LIFE CYCLE Bacteriophages are viruses that infect bacteria The structural genes of bacteriophages are often in an operon arrangement Their study has greatly advanced our basic knowledge of genetic regulation Like bacterial operons, phage operons can be controlled by repressor proteins or activator proteins To understand how this works, we will examine the two life cycles of phage l (lambda) Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-63 Life Cycles of Phage l Phage l can bind to the surface of a bacterium and inject its genetic material into the bacterial cytoplasm The phage will then proceed along only one of two alternative life cycles Lytic cycle Lysogenic cycle Let’s review Figure 6.9 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-64 This process is termed induction It will undergo the lytic cycle Prophage can exist in a dormant state for a long time Virulent phages only undergo a lytic cycle Figure 6.9 Temperate phages can follow both cycles Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-65 Figure 14.18 shows the genome of phage l Inside the viral head, phage l DNA is linear After injection into the bacterium, the two ends attach covalently to each other forming a circle The organization of the genes within this circular structure reflects the two alternative life cycles of the virus The genes in the top center are transcribed very soon after infection, at the beginning of either life cycle The pattern of their expression determines which of the two cycles prevails The genes on the left side of the viral genome encode proteins that are responsible for the lysogenic infection The genes on the right side of the viral genome encode proteins that are responsible for the lytic infection Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-66 Figure 14.18 14-67 Life Cycles of Phage l So how is the decision made between the lytic and lysogenic cycles? The choice depends on the actions of several genetic regulatory proteins The process is quite detailed It involves a series of intricate steps in which these proteins bind to several different sites in the l genome Refer to Figure 14.19 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-68 The N protein is an antiterminator Encoding two proteins: N and cro It binds to RNA polymerase and prevents transcriptional termination cIII gene encodes a protein that helps stabilize the cII activator protein Figure 14.19 cII gene encodes an activator protein The O and P genes encode enzymes needed to initiate DNA synthesis The Q gene encodes another antiterminator needed for the lytic cycle 14-69 cII/cIII activates transcription from PI and PRE PRE = Promoter for l Repressor during Establishment of lysogenic cycle int gene encodes the protein integrase, which integrates l DNA into the bacterial chromosome The l repressor binds to operators that are adjacent to PR and PL It thus inhibits the expression of genes required for the lytic cycle The l repressor also activates PRM This is sufficient to make enough l Repressor to Maintain the lysogenic cycle Figure 14.19 14-70 The cro protein binds to two operators OR and OL Binding to OL inhibits transcription from PL Binding to OR has several effects 1. It inhibits transcription from PRM in the leftward direction •This prevents the expression of the cI gene which encodes the l repressor 2. It allows a low level of transcription from PR in the rightward direction • This enables the transcription of the O, P and Q genes The O and P proteins are necessary for the replication of l DNA Figure 14.19 The Q protein is an antiterminator that permits transcription through another promoter, PR’ PR’ controls a very large operon that encodes the proteins necessary for the assembly of the phage coat, packaging of the DNA and lysis of the bacterial cell 14-71 Cellular Proteases Influence the Choice Between the Two Cycles The activity of the cII protein plays a key role in directing l to the lysogenic or lytic cycle The cII protein is easily degraded by cellular proteases produced by E. coli Whether or not these proteases are produced depends on the environmental conditions Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-72 If the growth conditions are very favorable, the intracellular levels of the proteases are high The cII protein tends to be degraded Instead, the cro protein slowly accumulates to high levels Therefore, PRE cannot be activated and the l repressor is not made The binding of the cro protein to OR prevents transcription of the l repressor from PRM At the same time, the cro protein allows the lytic cycle to proceed Thus, environmental conditions that are favorable for growth promote the lytic cycle This makes sense because a sufficient supply of nutrients is necessary to synthesize new bacteriophages Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-73 If the nutrients are limiting (starvation conditions), the cellular proteases are relatively inactive The cII protein builds up much more quickly than cro Therefore, the cII protein will turn on PRE The l repressor is made Thus, environmental conditions that are unfavorable for growth promote the lysogenic cycle This makes sense because there may not be sufficient nutrients for the production of new bacteriophages Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-74 After lysogeny is established, certain environmental conditions can also favor induction to the lytic cycle For example, exposure to UV light recA (a cellular protein normally involved in DNA recombination) detects the DNA damage It cleaves the l repressor and inactivates it This allows transcription from PR Therefore, the cro protein will accumulate It is activated to become a protease Favoring the lytic cycle This makes sense, because the exposure to UV light may have already damaged the bacterium to the point where further bacterial growth and division are prevented Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-75 The OR Region Provides a Genetic Switch Between the Two Cycles To understand how this switch works, we need to take a closer look at the OR region The OR region contains three operator sites, designated OR1, OR2, and OR3 These operator sites control two promoters, PR and PRM, which transcribe in opposite directions The l repressor protein or the cro protein can bind to any or all of the three operator sites This binding governs the switch between the lysogenic and the lytic cycles Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-76 The OR Region Provides a Genetic Switch Between the Two Cycles Two critical issues influence this binding 1. The relative affinities that the regulatory proteins have for these operator sites 2. The concentrations of these regulatory proteins in the cell Refer to Figure 14.20 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-77 Cro protein has the highest affinity to OR3 and simiar affinity to OR2 then OR1 l repressor has the highest affinity to OR1 then OR2 then OR3 l repressor is a dimer This binding inhibits transcription from PR So the lytic cycle is switched off l repressor falls off OR3 first Figure 14.20 cro protein is a dimer via cooperative interaction This binding blocks transcription from PRM So the lysogenic cycle is switched off PR is not needed in the later stages of the lytic cycle 14-78 Genetic switches, like the one just described in phage l, are also important in the developmental pathways of bacteria and eukaryotes For example The choice between sporulation and vegetative growth in bacteria Initiation of cell differentiation during development in eukaryotes Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-79