* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Host-induced epidemic spread of the cholera bacterium
RNA interference wikipedia , lookup
X-inactivation wikipedia , lookup
Human genome wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Heritability of IQ wikipedia , lookup
RNA silencing wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Public health genomics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Non-coding RNA wikipedia , lookup
Essential gene wikipedia , lookup
Oncogenomics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Metagenomics wikipedia , lookup
Microevolution wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Pathogenomics wikipedia , lookup
Genome evolution wikipedia , lookup
Gene expression programming wikipedia , lookup
Designer baby wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genome (book) wikipedia , lookup
Genomic imprinting wikipedia , lookup
Minimal genome wikipedia , lookup
Ridge (biology) wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
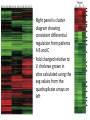
Host-induced epidemic spread of the cholera bacterium Theresa Graebener, Salomon Garcia, Claudia Campos Journal Club Presentation Biological Databases 367-01 October 19, 2010 Outline Background on Vibrio cholerae • • • • • It’s a gram negative bacterium Has polar flagellum Comma shaped Main cause for cholera V. cholerae has 2 circular chromosomes of unequal size that encode for an approximate of 3885 genes. The experiment itself… • For this study human subjects were gathered from the International Centre for Diarrhoeal Disease Research (ICDDR) • Subjects were from Dhaka, Bangladesh mainly due to bacterium being in it’s natural habitat • Serological analysis was done in order to identify if stool samples that were positive for V.cholerae 01 Inaba El Tor Bacterial Strain responsible for the spread of V.cholerae • Strain (DSM-V984) was strain that had been previously isolated and is marked by the deletion of the lacZ gene. It allows for the enumeration of in vitro and the stool samples • Strain (DSM-V984) was mixed with V.cholerae in stool • Mixture was injected to 3-5 day old Swiss Webster mice. • Mice were euthanized after 20-24hrs and bacteria was recovered Testing of the human shed V.cholerae occurred in order to test if the hyperinfectious phenotype was maintatined. • V.cholerae samples that were freshly shed were diluted in pond water that was free from contaminants of V.cholerae • Incubation at room temperature for 5 hrs, then diluted samples were mixed with in vitro grown competitor strains • This mixture was injected to mice • Hyperinfectious state remained the same the only difference being human passage of the bacteria enhances water bourne spread which lowers the dose to secondary individuals Transcriptional profiling of human shed V.cholerae • Spotted DNA microarray with 87% of known open reading frames • Samples of the 3 patients were taken then later frozen in order to prepare for total RNA. • RNA analyzed using agarose gel electrophoresis • Strain DSM-V999 from one of the 3 samples, RNA was isolated after in vitro growth Statistical Analysis reveals significant genes are responsible for gene expressions • 237 genes were differentially regulated • 44 genes were induced • 193 genes were repressed in human stool samples • Transcriptomes were similar to strain DSMV999, 3120 out of 3357 open reading frames that were examined were expressed Competition assays take advantage of concentration differences • On A samples mixed directly with in-vitro grown DSMV984 • On B samples incubated in local pond water for 5 hours • The Data points represent the output ratio of stoolderived V. cholerae to in-vitro grown competitor strain after correction for deviations in input rations. • Each data point represents output from a single animal • The geometric mean is indicated by horizontal bar • (n) is the total number of animals per experiment The transcription profiles were used to compare genetic similarities and differences The cluster diagram shows the wholegenome expression profile of V. cholerae recovered from the stools of three ICDDR patients • Patients labeled A B or C (in blue) • Replicate arrays are numbered 1-4 (in orange) • Right panel is cluster diagram showing consistent differential regulation from patients A B and C • Fold changed relative to V. cholerae grown in vitro calculated using the avg values from the quadruplicate arrays on left • Red indicated a minimum twofold increase in expression • Green represents a minimum twofold reduction in expression • Representative genes listed and their relative location indicated by arrow Differences found between human shed V.cholerae and strain DSM-V999 • Increased expression of genes required for biosynthesis of amino acids, iron uptake systems, ribosomal proteins, and formation of periplasmic nitrate reductase complex • V. cholerae moves from rich nutrient environment to poor environment which is purged. Several genes were found to be differentially expressed in stool samples • These genes were found to be factors for infection of mice and humans • VC0468 strain was found to play a significant role in the ability of V.cholerae to tolerate acid References Merrell D. Scott, Butler Susan M., QadriFirdausi, Dolganov Nadia A., Alam Ahsfaqui, Cohen Mitchell B., Calderwood Stephen B., Schoonik Gary K., & Camilli Andrew. Host-induced epidemic spread of the cholera bacterium. Nature. 2002. Vol, 417,