* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download NCBI Molecular Biology Resources
Whole genome sequencing wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Minimal genome wikipedia , lookup
Human genetic variation wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Copy-number variation wikipedia , lookup
Gene expression programming wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Transposable element wikipedia , lookup
Gene therapy wikipedia , lookup
Messenger RNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Public health genomics wikipedia , lookup
Gene desert wikipedia , lookup
Genetic engineering wikipedia , lookup
Non-coding DNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genome (book) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene nomenclature wikipedia , lookup
Primary transcript wikipedia , lookup
Epitranscriptome wikipedia , lookup
Human Genome Project wikipedia , lookup
Pathogenomics wikipedia , lookup
Point mutation wikipedia , lookup
Human genome wikipedia , lookup
Genomic library wikipedia , lookup
Microevolution wikipedia , lookup
Genome evolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Metagenomics wikipedia , lookup
Genome editing wikipedia , lookup
NCBI FieldGuide
NCBI Molecular Biology
Resources
A Field Guide
August 2-3, 2005
University of Massachusetts
• The NCBI Entrez System
• NCBI Sequence Databases
– Primary data: GenBank
– Derivative data: RefSeq, Gene, Genome
– Beyond Refseq: UniGene, Trace Archive
• NCBI Genomic Resources
** Intermission **
• BLAST
• Protein Structure and Function
• Sequence polymorphisms and phenotypes
NCBI FieldGuide
NCBI Resources
Bethesda, MD
NCBI FieldGuide
The National Institutes of Health
• Created as a part of NLM in 1988
–
–
–
–
Establish public databases
Perform research in computational biology
Develop software tools for sequence analysis
Disseminate biomedical information
NCBI FieldGuide
The National Center for
Biotechnology Information
Text
Entrez
Sequence
BLAST
Structure
VAST
NCBI FieldGuide
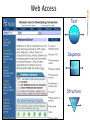
Web Access
600,000
NCBI FieldGuide
NCBI Web Traffic
User’s per day
World
Internet Users
500,000
400,000
US
Internet Users
300,000
200,000
100,000
1998
1999
2000
2001
2002
2003
2004
Christmas and New Year’s Day
2005
30,000 files per day
620 Gigabytes per day
NCBI FieldGuide
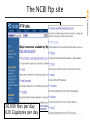
The NCBI ftp site
• NCBI accepts submissions of primary data
• NCBI develops tools to analyze these data
• NCBI uses these tools to create derivative
databases based on the primary data
• NCBI provides free search, link, and
retreival of these data, primarily through
the Entrez system
NCBI FieldGuide
What does NCBI do?
• Primary Databases
– Original submissions by experimentalists
– Content controlled by the submitter
• Examples: GenBank, SNP, GEO, PubChem Substance
• Derivative Databases
– Built from primary data
– Content controlled by third party (NCBI)
• Examples: Refseq, TPA, RefSNP, UniGene, Protein, Structure,
Conserved Domain, PubChem Compound
NCBI FieldGuide
Types of Databases
Algorithms
Sequencing
Centers
GenBank
Updated ONLY
by submitters
INV VRT PHG VRL
UniSTS
EST
STS
GSS
HTG
UniGene
NCBI FieldGuide
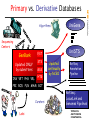
Primary vs. Derivative Databases
Updated
continually
by NCBI
RefSeq:
Annotation
Pipeline
PRI ROD PLN MAM BCT
Curators
Labs
RefSeq:
LocusLink and
Genomes Pipelines
TATAGCCG
AGCTCCGATA
CCGATGACAA
•
•
•
•
•
A system of 29 linked databases
A text search engine
A tool for finding biologically linked data
A retrieval engine
A virtual workspace for manipulating large
datasets
NCBI FieldGuide
What is Entrez?
NCBI FieldGuide
The Entrez System: Text Searches
• Each record is assigned a UID
– unique integer identifier for internal tracking
– GI number for Nucleotide
• Each record is given a Document Summary
– a summary of the record’s content (DocSum)
• Each record is assigned links to biologically
related UIDs
• Each record is indexed by data fields
– [author], [title], [organism], and many others
NCBI FieldGuide
Entrez Databases
The backbone of NCBI
[organism]
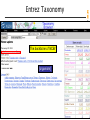
NCBI FieldGuide
Entrez Taxonomy
• GenBank: Primary Data (97.9%)
– original submissions by experimentalists
– submitters retain editorial control of records
– archival in nature
• RefSeq: Derivative Data (2.1%)
– curated by NCBI staff
– NCBI retains editorial control of records
– record content is updated continually
NCBI FieldGuide
An Entrez Database - Nucleotide
Primary Data
• DDBJ / EMBL / GenBank 56,865,268
Derivative Data
• RefSeq
• PDB
• Third Party Annotation
Total
1,226,084
5,973
4,650
58,101,975
NCBI FieldGuide
Entrez Nucleotide
What is GenBank?
•
•
•
•
Nucleotide only sequence database
Archival in nature
Each record is assigned a stable accession number
GenBank Data
– Direct submissions (traditional records )
– Batch submissions (EST, GSS, STS)
– ftp accounts (genome data)
• Three collaborating databases
– GenBank
– DNA Database of Japan (DDBJ)
– European Molecular Biology Laboratory (EMBL)
Database
NCBI FieldGuide
NCBI’s Primary Sequence Database
NIH
Sequin
BankIt
ftp
NCBI FieldGuide
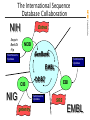
The International Sequence
Database Collaboration
Entrez
NCBI
GenBank
•Submissions
•Updates
•Submissions
•Updates
EMBL
CIB
NIG
DDBJ
•Submissions
•Updates
getentry
EBI
SRS
EMBL
Release 148
June 2005
45,236,251
49,398,852,122
>140,000
Records
Nucleotides
Species
172 Gigabytes
785 files
• full release every two months
• incremental and cumulative updates daily
• available only through internet
ftp://ftp.ncbi.nih.gov/genbank/
NCBI FieldGuide
GenBank Releases
NCBI FieldGuide
The Growth of GenBank
50
45
Basepairs
Records
Release 148:
35
25
35
45.2 million records
49.4 billion nucleotides
30
30
25
Average doubling time ≈ 14 months*
20
20
15
15
Date
Jun-04
Jun-02
Jun-00
Jun-98
Jun-96
Jun-94
0
Jun-92
0
Jun-90
5
Jun-88
5
Jun-86
10
Jun-84
10
Jun-82
Base Pairs (billions)
40
40
Records (millions)
45
PRI
ROD
PLN
BCT
INV
VRT
VRL
MAM
PHG
SYN
UNA
(28)
(14)
(13)
(10)
(7)
(7)
(4)
(2)
(1)
(1)
(1)
Primate
Rodent
Plant and Fungal
Bacterial/Archeal
Invertebrate
Other Vertebrate
Viral
Mammalian
Phage
Synthetic
Unannotated
EST
GSS
HTG
HTC
STS
(349)
(120)
(62)
(6)
(5)
Expressed Sequence Tag
Genome Survey Sequence
High Throughput Genomic
High Throughput cDNA
Sequence Tagged Site
Traditional
NCBI FieldGuide
GenBank Divisions
•Direct Submissions (Sequin/Bankit)
•Accurate (~1 error per 10,000 bp)
•Well characterized
•Organized by taxonomy
Bulk
•From sequencing projects
•Batch submissions (ftp/email)
•Inaccurate
•Poorly Characterized
•Organized by sequence type
LOCUS
DEFINITION
AY182241
1931 bp
mRNA
linear
PLN 04-MAY-2004
Malus x domestica (E,E)-alpha-farnesene synthase (AFS1) mRNA,
complete cds.
ACCESSION
AY182241
VERSION
AY182241.2 GI:32265057
KEYWORDS
.
SOURCE
Malus x domestica (cultivated apple)
ORGANISM Malus x domestica
Eukaryota; Viridiplantae; Streptophyta; Embryophyta; Tracheophyta;
Spermatophyta; Magnoliophyta; eudicotyledons; core eudicots;
rosids; eurosids I; Rosales; Rosaceae; Maloideae; Malus.
REFERENCE
1 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Cloning and functional expression of an (E,E)-alpha-farnesene
synthase cDNA from peel tissue of apple fruit
JOURNAL
Planta 219, 84-94 (2004)
REFERENCE
2 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Direct Submission
JOURNAL
Submitted (18-NOV-2002) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
REFERENCE
3 (bases 1 to 1931)
AUTHORS
Pechous,S.W. and Whitaker,B.D.
TITLE
Direct Submission
JOURNAL
Submitted (25-JUN-2003) PSI-Produce Quality and Safety Lab,
USDA-ARS, 10300 Baltimore Ave. Bldg. 002, Rm. 205, Beltsville, MD
20705, USA
REMARK
Sequence update by submitter
COMMENT
On Jun 26, 2003 this sequence version replaced gi:27804758.
FEATURES
Location/Qualifiers
source
1..1931
/organism="Malus x domestica"
/mol_type="mRNA"
/cultivar="'Law Rome'"
/db_xref="taxon:3750"
/tissue_type="peel"
gene
1..1931
/gene="AFS1"
CDS
54..1784
/gene="AFS1"
/note="terpene synthase"
/codon_start=1
/product="(E,E)-alpha-farnesene synthase"
/protein_id="AAO22848.2"
/db_xref="GI:32265058"
/translation="MEFRVHLQADNEQKIFQNQMKPEPEASYLINQRRSANYKPNIWK
NDFLDQSLISKYDGDEYRKLSEKLIEEVKIYISAETMDLVAKLELIDSVRKLGLANLF
EKEIKEALDSIAAIESDNLGTRDDLYGTALHFKILRQHGYKVSQDIFGRFMDEKGTLE
NHHFAHLKGMLELFEASNLGFEGEDILDEAKASLTLALRDSGHICYPDSNLSRDVVHS
LELPSHRRVQWFDVKWQINAYEKDICRVNATLLELAKLNFNVVQAQLQKNLREASRWW
ANLGIADNLKFARDRLVECFACAVGVAFEPEHSSFRICLTKVINLVLIIDDVYDIYGS
EEELKHFTNAVDRWDSRETEQLPECMKMCFQVLYNTTCEIAREIEEENGWNQVLPQLT
KVWADFCKALLVEAEWYNKSHIPTLEEYLRNGCISSSVSVLLVHSFFSITHEGTKEMA
DFLHKNEDLLYNISLIVRLNNDLGTSAAEQERGDSPSSIVCYMREVNASEETARKNIK
GMIDNAWKKVNGKCFTTNQVPFLSSFMNNATNMARVAHSLYKDGDGFGDQEKGPRTHI
LSLLFQPLVN"
ORIGIN
1 ttcttgtatc ccaaacatct cgagcttctt gtacaccaaa ttaggtattc actatggaat
61 tcagagttca cttgcaagct gataatgagc agaaaatttt tcaaaaccag atgaaacccg
121 aacctgaagc ctcttacttg attaatcaaa gacggtctgc aaattacaag ccaaatattt
181 ggaagaacga tttcctagat caatctctta tcagcaaata cgatggagat gagtatcgga
241 agctgtctga gaagttaata gaagaagtta agatttatat atctgctgaa acaatggatt
1801 aataaatagc agcaaaagtt tgcggttcag ttcgtcatgg ataaattaat ctttacagtt
1861 tgtaacgttg ttgccaaaga ttatgaataa aaagttgtag tttgtcgttt aaaaaaaaaa
1921 aaaaaaaaaa a
//
The Flatfile Format
Header
Feature Table
Sequence
NCBI FieldGuide
A Traditional GenBank Record
Indexing for Nucleotide UID 4680720
Field
[primary accession]
[title]
[organism]
[sequence length]
[modification date]
[properties]
Indexed Terms
M17755
Homo sapiens thyroid peroxidase (TPO) mRNA…
Homo sapiens
3060
1999/04/26
biomol mrna
gbdiv pri
srcdb genbank
NCBI FieldGuide
An Example Record – M17755
NCBI FieldGuide
M17755: Feature Table
TPO [gene name]
CDS position in bp
thyroiditis
[text word]
thyroid peroxidase
[protein name]
protein
accession
The sequence itself
is not indexed…
Use BLAST for that!
NCBI FieldGuide
Sequence: 99.99% Accurate
•
•
•
•
•
•
•
GenPept (DDBJ, EMBL, GenBank)
RefSeq
PIR
Swiss Prot
PDB
PRF
Third Party Annotation
Total
4,444,405
1,753,167
222,395
189,005
68,621
12,079
4,219
6,693,891
NCBI FieldGuide
Entrez Protein
PIR
RefSeq
no mRNA!
NM_000537
SWISS-PROT
GenPept
no mRNA!
M17755
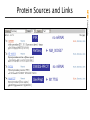
NCBI FieldGuide
Protein Sources and Links
First seen at NCBI, not
first seen at GenBank!
Version and GI change only if the sequence changes
The accession number always retrieves the most recent version
NCBI FieldGuide
Sequence Revisions
NCBI FieldGuide
Update without a Sequence Change
June 15, 1989!
GenBank came
to NCBI in 1992!
NCBI FieldGuide
Update with a Sequence Change
ASN.1 – The Raw Data
flat file
XML (4 flavors)
FASTA
NCBI FieldGuide
GenBank File Formats
/************************************************************************
*
*
asn2ff.c
*
convert an ASN.1 entry to flat file format, using the FFPrintArray.
*
**************************************************************************/
#include <accentr.h>
#include "asn2ff.h"
#include "asn2ffp.h"
#include "ffprint.h"
#include <subutil.h>
#include <objall.h>
#include <objcode.h>
#include <lsqfetch.h>
#include <explore.h>
Toolbox Sources
ftp> open ftp.ncbi.nih.gov
.
.
#ifdef ENABLE_ID1
ftp> cd toolbox
#include <accid1.h>
#endif
ftp> cd ncbi_tools
FILE *fpl;
Args myargs[] = {
{"Filename for asn.1 input","stdin",NULL,NULL,TRUE,'a',ARG_FILE_IN,0.0,0,NULL},
{"Input is a Seq-entry","F", NULL ,NULL ,TRUE,'e',ARG_BOOLEAN,0.0,0,NULL},
{"Input asnfile in binary mode","F",NULL,NULL,TRUE,'b',ARG_BOOLEAN,0.0,0,NULL},
{"Output Filename","stdout", NULL,NULL,TRUE,'o',ARG_FILE_OUT,0.0,0,NULL},
{"Show Sequence?","T", NULL ,NULL ,TRUE,'h',ARG_BOOLEAN,0.0,0,NULL},
ftp://ftp.ncbi.nlm.gov/toolbox/ncbi_tools
NCBI FieldGuide
NCBI Toolbox
term1 term2
If no [limit] is specified…
Organism? [ organism ]
Journal? [ journal ]
User compounds? search as phrase
Author? [author]
else [All Fields]
term1[limit] OP term2[limit] OP …
where
limit = Entrez indexing field (organism, author, …)
op = AND, OR, NOT
NCBI FieldGuide
Text Searches in Entrez
Limits
Provides a simple form for applying commonly used Entrez limits
Preview/Index
Allows access to the full indexing of each Entrez database
and aids in constructing complex queries
History
Provides access to previous searches in the current Entrez database
Clipboard
A temporary storage area for selected records
Details
Displays the detailed parsing of the current Entrez query, and
lists errors and terms without matches
NCBI FieldGuide
Entrez Tabs
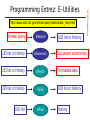
http://www.ncbi.nih.gov/entrez/query/static/eutils_help.html
Entrez query
ESearch
UID list or History
UID list or History
ESummary
UID list or History
EFetch
UID list or History
ELink
UID list or History
UID list
EPost
History
Document summaries
Formatted data
NCBI FieldGuide
Programming Entrez: E-Utilities
• Search Entrez Nucleotide
– 97.9% GenBank (primary data)
– 2.1% RefSeq (curated data)
Possible queries we’ve seen so far…
M17755 [primary accession]
thyroid peroxidase [title]
Homo sapiens [organism]
3060 [sequence length]
biomol mrna [properties]
srcdb genbank [properties]
TPO [gene name]
thyroiditis [text word]
thyroid peroxidase [protein name]
1999/04/26 [modification date]
gbdiv pri [properties]
NCBI FieldGuide
Finding Primary Sequences
Find nucleotide records for human thyroid peroxidase
human thyroid peroxidase
309 records
(("Homo sapiens“[Organism] OR human[All Fields]) AND
thyroid peroxidase[All Fields])
Field Limit!
human[organism] AND thyroid peroxidase
298 records
("Homo sapiens“[Organism] AND thyroid peroxidase[All Fields])
11 records aren’t human sequences!!
NCBI FieldGuide
A Starting Query
Entrez Nucleotide
GenBank
RefSeq
srcdb ddbj/embl/genbank[properties]
NCBI FieldGuide
Limit by Title and Database
srcdb refseq[properties]
#1: thyroid peroxidase AND human[orgn]
#2: thyroid peroxidase[title] AND human[orgn]
#3: #2 AND srcdb refseq[properties]
#4: #2 AND srcdb ddbj/embl/genbank[properties]
primary data
298
169
5
164
EST Division
Primate Division
#1:
#2:
#3:
#4:
NCBI FieldGuide
Limit by Genbank Division
gbdiv est[prop]
gbdiv pri[prop]
thyroid peroxidase AND human[orgn]
thyroid peroxidase[title] AND human[orgn]
#2 AND srcdb refseq[properties]
#2 AND srcdb ddbj/embl/genbank[properties]
#5: #4 AND gbdiv est[prop]
#6: #4 AND gbdiv pri[prop]
20
144
traditional GenBank records
298
169
5
164
Genomic DNA
cDNA
#1:
#2:
#3:
#4:
#5:
#6:
biomol genomic[prop]
biomol mrna[prop]
thyroid peroxidase AND human[orgn]
298
thyroid peroxidase[title] AND human[orgn]
169
#2 AND srcdb refseq[properties]
5
#2 AND srcdb ddbj/embl/genbank[properties] 164
#2 AND gbdiv est[prop]
20
#2 AND gbdiv pri[prop]
144
genomic DNA
#7: #6 AND biomol genomic[prop]
#8: #6 AND biomol mrna[prop]
mRNA / cDNA
26
118
NCBI FieldGuide
Limit by Biomolecule Type
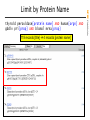
thyroid peroxidase[protein name] AND human[orgn] AND
gbdiv pri[prop] AND biomol mrna[prop]
118 records [title] 4 records [protein name]
NCBI FieldGuide
Limit by Protein Name
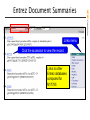
Links menu
Click the accession to view the record
Links to other
Entrez databases
computed for
M17755
NCBI FieldGuide
Entrez Document Summaries
Gene annotation based on M17755
Full text online articles about M17755
All polymorphisms in the TPO gene
DNA/RNA sequences similar to M17755
Graphical view of TPO gene annotation
Human phenotypes involving TPO
Microarray datasets for M17755
Protein translation of M17755
Literature abstracts about M17755
Sequence polymorphisms in M17755
Source organism of M17755
STS markers in the TPO gene
TPO links beyond NCBI
NCBI FieldGuide
Entrez Links for GI 4680720
NCBI FieldGuide
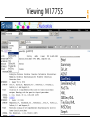
Viewing M17755
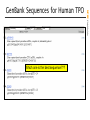
Which one is the best sequence???
NCBI FieldGuide
GenBank Sequences for Human TPO
NCBI’s Derivative Sequence Database
RefSeq Benefits
•
•
•
•
•
•
•
NCBI FieldGuide
RefSeq:
Non-redundant
Explicitly linked nucleotide and protein sequences
Updated to reflect current sequence data and biology
Validated by hand
Format consistency
Distinct accession series
Stewardship by NCBI staff and collaborators
ftp://ftp.ncbi.nih.gov/refseq/release
NCBI’s Derivative Sequence Database
• Curated transcripts and proteins
– NM_123456 NP_123456
– NR_123456 (non-coding RNA)
• Model transcripts and proteins
– XM_123456 XP_123456
– XR_123456 (non-coding RNA)
Nucleotide
Protein
• Assembled Genomic Regions (contigs)
– NT_123456 (BAC clones)
– NW_123456 (WGS)
• Other Genomic Sequence
– NG_123456 (complex regions, pseudogenes)
– NZ_ABCD12345678 (WGS) ZP_123456
• Chromosome records in Entrez Genome
– NC_123456 (chromosome; microbial or organelle genome)
NCBI FieldGuide
RefSeq:
Genome annotation
Longest mRNA
NMs must have
cDNA support
NCBI FieldGuide
Creating NM Records
NM_000547: variant 1
COMMENT REVIEWED REFSEQ: This record has been curated by NCBI staff.
The reference sequence was derived from M17755.2 and AW874082.1.
On Feb 25, 2003 this sequence version replaced gi:21361188.
NM_175719: variant 2
EST that completes 3’ end
COMMENT REVIEWED REFSEQ: This record has been curated by NCBI staff.
The reference sequence was derived from J02970.1, AW874082.1 and M17755.2.
Nucleotide
Protein
NCBI FieldGuide
NM/NP Records in Entrez
Genomic DNA
(NC, NT, NW)
Scanning....
Model mRNA (XM)
(XR)
Curated mRNA (NM)
(NR)
RefSeq
Genbank
Sequences
NCBI FieldGuide
Annotating the Gene
Model protein (XP)
= ?!
Curated Protein (NP)
GenBank
RefSeq
Gene
Nucleotide
• Entrez Gene is the central depository for information about a gene
available at NCBI, and often provides links to sites beyond NCBI
• Entrez Gene includes records for organisms that have NCBI Reference
Sequences (RefSeqs)
• Entrez Gene records contain RefSeq mRNAs, proteins, and genomic
DNA (if known) for a gene locus, plus links to other Entrez databases
• NCBI RefSeqs are based on primary sequence data in GenBank
NCBI FieldGuide
Entrez Gene and RefSeq
NCBI FieldGuide
Entrez Gene: RefSeq Annotations
NCBI FieldGuide
NM/NP Records in Entrez Gene
NM
NCBI FieldGuide
Entrez Gene RefSeq Graphics
NP
Entrez Gene
NCBI FieldGuide
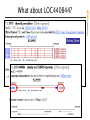
What about LOC440844?
Is there any GenBank support for this mRNA?
srcdb ddbj/embl/genbank[prop] AND biomol mrna[prop]
no full-length hit
NCBI FieldGuide
BLAST Results for XM_496543
XM records are models based only on genomic sequence, and are subject
to revision or removal with each new build of that genome.
BLAST the XM against the RefSeq database to look for a replacement:
Query= gi|20850420|ref|XM_124429.1|
Mus musculus expressed sequence AA553001 (AA553001), mRNA
gi|19527087|ref|NM_133873.1|
Mus musculus DNA segment, Chr 4, Wayne State University 114,
expressed (D4Wsu114e), mRNA Length=1898
Score = 3701.55 bits (1867), Expect = 0
Identities = 1870/1871 (99%), Gaps = 0/1871 (0%) Strand=Plus/Plus
NCBI FieldGuide
The Perils of the XM
Bos taurus:
37541
Oryza sativa (japonica cultivar-group):
36836
Danio rerio:
30577
Homo sapiens:
29261
Arabidopsis thaliana:
28953
Mus musculus:
27033
Rattus norvegicus:
23975
Pan troglodytes:
21810
Caenorhabditis elegans:
21124
Drosophila melanogaster:
19412
Aspergillus nidulans FGSC A4:
18951
Gallus gallus:
18120
Canis familiaris:
16891
Anopheles gambiae str. PEST:
15328
Plasmodium chabaudi:
14747
Candida albicans SC5314:
13672
Dictyostelium discoideum:
13570
Ustilago maydis 521:
13044
Plasmodium berghei:
11778
Gibberella zeae PH-1:
11640
Magnaporthe grisea 70-15:
11109
Neurospora crassa:
10079
Aspergillus fumigatus Af293:
9923
Entamoeba histolytica HM-1:IMSS:
9772
Cryptococcus neoformans var. neoformans JEC21: 6594
NCBI FieldGuide
Eukaryotic NM/XM Records
Giardia lamblia ATCC 50803:
6569
Yarrowia lipolytica CLIB99:
6521
Debaryomyces hansenii CBS767:
6318
Apis mellifera:
6292
Kluyveromyces lactis NRRL Y-1140: 5327
Candida glabrata CBS138:
5181
Schizosaccharomyces pombe 972h-: 5035
Eremothecium gossypii:
4718
Theileria parva:
4079
Xenopus tropicalis:
4069
Cryptosporidium hominis:
3886
Cryptosporidium parvum:
3396
Sus scrofa:
938
Trypanosoma brucei:
599
Ovis aries:
253
Strongylocentrotus purpuratus:
215
Felis catus:
162
Plasmodium yoelii yoelii:
105
Takifugu rubripes:
7
Ciona intestinalis:
3
Trypanosoma cruzi:
3
GenBank Components
(clones, WGS)
NT/NW Contigs
NC
Genome
Assembly
NM/XM
Master
mRNA
Components
Components
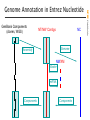
NCBI FieldGuide
Genome Annotation in Entrez Nucleotide
curated mRNA
genomic contig on human chromosome 2
containing NM_000547
human chromosome 2
the 21 contigs of the
chromosome 2 assembly
NCBI FieldGuide
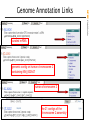
Genome Annotation Links
Genomic sequence
NCBI FieldGuide
Getting the Annotation Details
ACCESSION NC_000002 REGION: 1396242..1525502
ACCESSION NC_000002 REGION: 1396242..1525502
exon-intron structure
These flat files contain all annotations in the gene and the full, explicit sequence
NCBI FieldGuide
Getting the Annotation Details
Gene symbol: human thyroid peroxidase (TPO)
tpo [sym] AND human [organism]
NCBI FieldGuide
Searching Entrez Gene
Protein name: topoisomerase genes from Archaea
topoisomerase[gene/protein name] AND archaea [organism]
Chromosome and Links: genes on human chromosome 2 with OMIM links
2 [chromosome] AND gene omim [filter] AND human [organism]
RefSeq status and variants: Reviewed RefSeqs with transcript variants
srcdb refseq reviewed[prop] AND has transcript variants[prop]
Disease and Gene Ontology: Membrane proteins linked to cancer
integral to plasma membrane[gene ontology] AND cancer [dis]
Microarray datasets for TPO
NCBI FieldGuide
Gene Links in Entrez
Gene homologs for TPO
DNA and RNA sequences for TPO
Phenotypes involving TPO
Protein sequences for TPO
Literature abstracts about TPO
Sequence polymorphisms in TPO
Species whose genome has this TPO gene
STS markers in the TPO gene
ESTs aligned to the TPO gene
NCBI now accepts the submission of new annotations
of existing GenBank sequences.
NCBI FieldGuide
Third Party Annotation
(TPA) Database
• Submissions must be published in a peer-reviewed journal.
• Facilitates the annotation of sequences by experts.
Examples of sequences appropriate for TPA are:
Annotation of features on gene and/or mRNA sequences
Assembled “full length” genes and/or mRNAs
What should not be submitted to TPA?
Synthetic constructs (such as cloning vectors) that use well-characterized,
publicly available genes, promoters, or terminators
Updates or changes to existing sequence data
Sequence annotations without experimental evidence
If your organism does not have RefSeqs…
• UniGene : gene-based clusters of cDNAs and ESTs
• WGS sequences in Entrez Nucleotide (wgs[prop])
• Trace Archive
NCBI FieldGuide
Beyond RefSeq
A gene-oriented view of sequence entries
•MegaBlast based automated sequence clustering
•Now informed by genome hits New!
•Nonredundant set of gene oriented clusters
•Each cluster a unique gene
•Information on tissue types and map locations
•Includes known genes and uncharacterized ESTs
•Useful for gene discovery and selection of
mapping reagents
NCBI FieldGuide
What is UniGene?
Top Ten
1. Human
2. Rice
3. Mouse
4. Cow
5. Wheat
6. Zebrafish
7. Pig
8. Chicken
9. Frog (X. laevis)
10. Frog (X. tropicalis)
NCBI FieldGuide
Organisms in UniGene
by link
by Entrez search
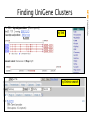
NCBI FieldGuide
Finding UniGene Clusters
NCBI FieldGuide
UniGene Cluster for TPO
GPL
Platform
descriptions
GSM
GSE
Grouping of
Raw/processed
slide/chip data
spot intensities
from a single “a single experiment”
slide/chip
Entrez GEO
Curated by
NCBI
NCBI FieldGuide
Submitted by
Manufacturer*
Submitted by
Experimentalists
GDS
Grouping of
experiments
Entrez
GEO Datasets
NCBI FieldGuide
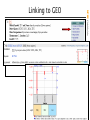
Linking to GEO
NCBI FieldGuide
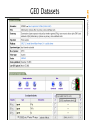
GEO Datasets
• Traditional GenBank Divisions
• 300 + projects
–
–
–
–
–
Viruses
Bacteria
Environmental sequences
Archaea
73 Eukaryotes featuring:
•
•
•
•
•
•
Cow, Chicken, Rat, Mouse, Dog, Chimpanzee, Human
Pufferfish (2), Zebrafish
Honeybee, Anopheles, Fruit Flies (4), Silkworm
Nematode (C. briggsae)
Yeasts (9), Aspergillus (3)
Rice
NCBI FieldGuide
Whole Genome Shotgun Projects
NCBI FieldGuide
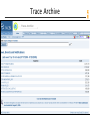
Trace Archive
NCBI FieldGuide
Short-tailed opossum traces
All are RefSeq NC records in Entrez Genome
• Full chromosomal
sequences are provided
• Genes are annotated
• The annotation can be
shown graphically and
linked to sequence records
NCBI FieldGuide
Viewing Simple Genomes
NCBI FieldGuide
NCBI FieldGuide
mutL
NCBI Map Viewer
• Map Viewer Home Page
– Shows all supported organisms
– Provides links to genomic BLAST
• Genome Overview Page
– Provides links to individual chromosomes
– Shows hits on a genome graphically
• Chromosome Viewing Page
– Allows interactive views of annotation details
– Provides numerous maps unique to each genome
NCBI FieldGuide
Viewing Complex Genomes
NCBI FieldGuide
Map Viewer Home Page
Search the maps
Genomic BLAST
Species-specific help!
NCBI FieldGuide
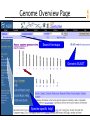
Genome Overview Page
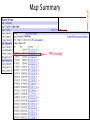
Map Summary
Add or remove maps
Master Map
with exploded content
Genes
UniGene
Contigs
Zooming
Controls
Ideogram
NCBI FieldGuide
Chromosome Viewing Page
TPO’s contig!
NCBI FieldGuide
Map Summary
Map content varies greatly by species!
• Sequence Maps
• Core assembly
• Annotation evidence
• Clones & Markers
• Polymorphisms
• Links & Features
• Genetic Maps
• Cytogenetic maps
• Linkage maps
• Radiation hybrid maps
Assembly
Contig
Component
Transcript
Gene
NCBI FieldGuide
Map Content
NCBI FieldGuide
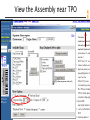
View the Assembly near TPO
NT_033000
1255072
1563756
NCBI FieldGuide
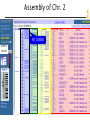
Assembly of Chr. 2
NCBI FieldGuide
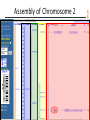
Assembly of Chromosome 2
NCBI FieldGuide
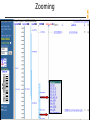
Zooming
NCBI FieldGuide
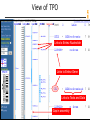
View of TPO
Links to Entrez Nucleotide
Links to Entrez Gene
Links to Tools and Data
Gap in assembly
Map content varies greatly by species!
• Sequence Maps
• Core assembly
• Annotation evidence
• Clones & Markers
• Polymorphisms
• Links & Features
• Genetic Maps
• Cytogenetic maps
• Linkage maps
• Radiation hybrid maps
Ab initio (model)
GenBank DNA
EST
UniGene
Gene
NCBI FieldGuide
Map Content
GenBank records not used in
assembly
Aligned ESTs
NCBI FieldGuide
Annotation Evidence
UniGene Clusters
Ab initio models
Homologs by protein BLAST
NCBI FieldGuide
Entrez Homologene