* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download knockdown
Nucleic acid analogue wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Ridge (biology) wikipedia , lookup
Gene desert wikipedia , lookup
Community fingerprinting wikipedia , lookup
X-inactivation wikipedia , lookup
Genomic imprinting wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Polyadenylation wikipedia , lookup
List of types of proteins wikipedia , lookup
Molecular evolution wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome evolution wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Messenger RNA wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene regulatory network wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Non-coding RNA wikipedia , lookup
Gene expression wikipedia , lookup
Epitranscriptome wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
RNA interference in
specific gene silencing
('knockdown')
Christopher V. Jones
Jason Carter
RNA Interference
mRNA transcribed from DNA encodes for a protein
expressed by a certain gene
The presence of certain double-stranded RNA
(dsRNA) interferes with expression of a gene by
interfering w/ the translation of its mRNA
dsRNAs direct the creation of small interfering RNAs
(siRNAs) which target RNA-degrading enzymes
(RNAses) to destroy mRNA transcripts
complementary to the siRNAs
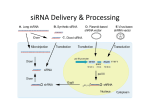
Small interfering RNA (siRNA)
dsRNA (usually 21-nt) with 2-nt overhangs on
either end, including a 5' phosphate group and a 3'
hydroxy (-OH) group
dsRNA enters RNAi pathway via enzyme Dicer
producing siRNA
siRNA molecules associate with a group of proteins
termed the RNA-induced silencing complex (RISC),
and directs the RISC to the target mRNA
Applications
Typically, a single mRNA translates about 5,000
protein copies
RNAi can be used experimentally to "knockout"
genes in organisms to help determine gene function
dsRNAs that trigger RNAi may be usable as drugs to
treat genetic disorders or cancers
dsRNA can repress essential genes in pathogens or
viruses that are dissimilar from any host genes
Advantages
Broad Applicability — Diseases for which abnormal
Therapeutic Precision — Side effects associated with
Target RNA Destruction — Most drugs only
gene function is a cause or a contributing factor are
potentially treatable with RNA interference
traditional drugs may be reduced or avoided by using RNAibased drugs designed to inhibit expression of only a targeted
gene and no others
temporarily prevent targeted protein function, RNAi-based
drugs are designed to destroy the target RNA stopping
undesirable protein production required for disease
progression
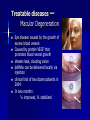
Treatable diseases —
Macular Degeneration
Eye disease caused by the growth of
excess blood vessels
Caused by protein VEGF that
promotes blood vessel growth
Vessels leak, clouding vision
dsRNAs can be delivered locally via
injection
clinical trial of two dozen patients in
2004
In two months:
¼ improved, ¾ stabilized
Treatable diseases —
HIV
In 2002, scientists at MIT accounted they could
interrupt various steps in the HIV life cycle using
RNAi in cell cultures
Mutates and evolves resistance too rapidly for any
single target mRNA
Molecular biologists at Colorado State University
have engineered RNAi therapy aiming at multiple
HIV genes
Clinical trials may start as early as 2006
Treatable diseases —
Cancer
involves mutant genes that promote uncontrolled
cell growth
researchers have silenced more than a dozen
known cancer-causing genes with RNAi in cell
cultures
delivery poses the key challenge for RNAi therapies:
how to reach and penetrate tumors
Could stop production of P-glycoprotein which
purges existing chemotherapy drugs from tumors,
thus enhancing existing treatments
siRNA Prediction
Given a target gene, how to design an siRNA to knock
it down?
Select a candidate subsequence from the target
gene
Not all subsequences are recognizable by Dicer
Arbitrary subsequence may knockdown unrelated
gene(s)
Identify siRNA patterns that are effective through
experimentation
Search entire genome to eliminate subsequences
with off-target specificity
siRNA Prediction Method from:
siDirect: highly effective target-specific siRNA design
software for mammalian RNA interference,
(Naito, Yamada, Ui-Tei, Morishita, Saigo, 2004)
Studies of several genes led to these heuristics:
A/U at the 5' end of the antisense strand
G/C at the 5' end of the sense strand
AU richness in the 5' terminal 1/3rd of the antisense strand
the absence of any G/C stretch exceeding 9 bp in length
siRNA Prediction Method from:
Rational siRNA design for RNA interference
(Reynolds, Leake, Boese, Scaringe, Marshall,
Khvorova, 2004)
At least 7 points are required to be scored as effective
siRNA
30%-52% GC content – Add 1 point
Three or more A/Us at positions 15-19 (sense) - Add 1
point for each A/U for a total up to 5 points. At least 3
points are required.
A at position 19 (sense) - Add 1 point
A at position 3 (sense) - Add 1 point
U at position 10 (sense) - Add 1 point
No G/C at position 19 (sense) - Subtract 1 point for not
satisfying this criterion.
No G at position 13 (sense) - Subtract 1 point for not
satisfying this criterion.
Filtering out off-target hits
Once we have predicted potentially effective
candidate siRNAs, we must search the entire
genome for off-target matches
Exhaustive search is expensive, but accurate:
Smith-Waterman algorithm
Approximate search: BLAST algorithm
Genes have introns that are spliced out of the
mRNA
Alternative-splicing means exons are spliced several
ways – we must search these areas also
Exhaustive vs. Approximate search
The human genome contains ~3B nt
Only 1.5% encodes proteins as genes
Must search ~45M nt, exon overlap sites, and alternative exon
overlaps
Must repeat search for each candidate siRNA
Exhaustive search is O(nm) time and space complexity
Smith-Waterman is a dynamic algorithm that finds optimal
local alignment using a scoring system, a substitution matrix,
and gap-scoring
Approximate search BLAST can run ~50 times faster using
heuristic approach
Approximate Search Basic Local Alignment Search Tool
BLAST breaks a search into stages
Searches for short matches of fixed length W between
query and database
If there is a matching word W, performs an ungapped
alignment between the query and database sequence,
extending the match in each direction
High-scoring matches then subjected to a gapped
alignment between the query sequence and the
database sequence using a variation of the SmithWaterman algorithm
Statistically significant matches are returned
Potential matches may get discarded due to heuristics
siRNA specificity
siRNA matches to any other gene of as few as 11
residues can lead to off-target silencing
High specificity has been observed with siRNAs that
have at least 3 mismatches to all other genes
Would be considered to have a mismatch tolerance
of 3
Higher mismatch tolerance indicates higher
specificity
Provides means to rank resulting siRNA candidates
for study
Conclusions
Hundreds of successful experiments in cell cultures,
and dozens in lab animals
siRNA delivery methods major hurdle
siRNA design will mature through competing
prediction heuristics and better characterization of
the RNAi machinery
As RNAi databases mature, novel biocomputing
approaches are likely
Optimistic many RNAi therapies will enter clinical
trials in next five years
Possible FDA approvals within the next decade
WebTools
siDirect:
http://design.rnai.jp/
Whitehead Institute siRNA:
http://jura.wi.mit.edu/bioc/siRNAext/
Wistar Bioinformatics Gene-specific siRNA selector:
http://bioinfo.wistar.upenn.edu/siRNA/siRNA.htm
Ambion siRNA design and databases:
http://www.ambion.com/techlib/misc/siRNA_tools.html
Web RNAi databases
http://www.rnainterference.org/
http://nematoda.bio.nyu.edu/cgi-bin/rnai/index.cgi
Bibliography
Review: Gene Silencing in mammals by small interfering RNAs, (McManus,
Sharp) Genetics Vol. 3 Oct. 2002, 737-747
Rational siRNA design for RNA interference (Reynolds, Leake, Boese,
Scaringe, Marshall, Khvorova) Nature Biotechnology Vol. 22:3 Mar. 2004, 326330.
siDirect: highly effective target-specific siRNA design software for mammalian
RNA interference, (Naito, Yamada, Ui-Tei, Morishita, Saigo) Nucleic Acids
Research Vol. 32 2004, 124-129.
Guidelines for the selection of highly effective siRNA sequences for
mammalian and chick RNA interference, (Ui-Tei, Naito, Takahashi, Haraguchi,
Okhi-Hamazaki, Juni, Ueda, Saigo, 2004) Nucleic Acids Research Vol. 32:3
2004
Potent and Persistent in-vivo anti-HBV activity of chemically modified siRNAs,
(Morrisey, Lockridge, et. al.) Nature Biotechnology July 2004