* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Bacterial Gene Expression—Lac Operon
Epigenetics of depression wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Designer baby wikipedia , lookup
Gene expression profiling wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression programming wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Messenger RNA wikipedia , lookup
Long non-coding RNA wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Microevolution wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Transcription factor wikipedia , lookup
Non-coding RNA wikipedia , lookup
Point mutation wikipedia , lookup
Epitranscriptome wikipedia , lookup
Primary transcript wikipedia , lookup
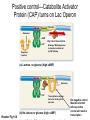
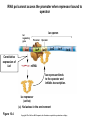
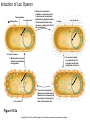
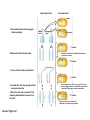
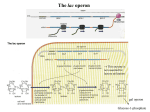
Study Guide/Outline—Bacterial Gene Regulation Bacterial Gene Regulation • What is an operon? How is it different from a eukaryotic gene? • In the lac operon, what cellular or environmental conditions must exist in order for the (WT) lac operon to express its genes? How do these environmental conditions positively or negatively regulate the operon? • What are the different parts, and their functions, of the operon? • How do mutations in “upstream” parts of the operon (promoter, operator, coding genes) affect the “downstream” areas of the operon? How do missense and nonsense mutations have different results? • The lacI gene is not part of the Lac Operon. How is the lac I gene involved with the Lac operon? • What kinds of mutations are cis-dominant? Trans-dominant? Constitutive ON? Constitutive-OFF? • How can a bacteria be a partial diploid? How does being diploid for the LacI gene create complexities in the regulation of the Lac Operon? Functions of lactose permease and b-galactosidase Lactose H+ Lactose permease H+ Cytoplasm CH2OH CH2OH O O OH HO H H Lactose H O OH H OH H H H H H OH H β-galactosidase side reaction O HO b-galactosidase H CH2OH O H OH H O CH2 O OH H H HO H OH CH2OH CH2OH H OH H O OH O OH H H H + b-galactosidase OH H OH H H H H HO H Allolactose H OH HO H OH Galactose H OH Glucose Brooker Fig 16.3b Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display Positive control—Catabolite Activator Protein (CAP) turns on Lac Operon CAP site Promoter Operator Allolactose cAMP High rate of transcription High rate of transcription CAP Repressor (inactive) Binding of RNA polymerase to promoter is enhanced by CAP binding. (a) Lactose, no glucose (high cAMP) CAP site Promoter Operator Repressor cAMP CAP Brooker Fig 16.8 Transcription is very low due to the binding of the repressor. (b) No lactose or glucose (high cAMP) But negative control Must be removed before positive control will result in transcription In absence of cAMP, transcription is very low (or hardly at all) CAP site Promoter Operator Allolactose CAP Repressor (inactive) Transcription rate is low due to the lack of CAP binding. (Inactive) (c) Lactose and glucose (low cAMP) CAP site Promoter Operator CAP (Inactive) Transcription is very low due to the lack of CAP binding and the binding of the repressor. (d) Glucose, no lactose (low cAMP) Brooker, Fig 16.8 RNA pol cannot access the promoter when repressor bound to operator lac regulatory gene lac operon Promoter lacI Constitutive expression of lacI lacP Operator lacO lacZ lacY lacA mRNA lac repressor binds to the operator and inhibits transcription. lac repressor (active) (a) No lactose in the environment Figure 16.4 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display (b) Lactose present Lactose causes repressor to fall off Operator Site RNA polymerase lacI lacP lacO lacZ lacY lacA Transcription mRNA Polycistronic mRNA b-galactosidase Lactose permease Galactoside transacetylase Allolactose Conformational change The binding of allolactose causes a conformational change that prevents the lac repressor from binding to the operator site. Figure 16.4 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display Induction of Lac Operon Transacetylase Lac repressor b-galactosidase 1. When lactose becomes available, a small amount of it is taken up and converted to allolactose by β-galactosidase. The allolactose binds to the repressor, causing it to fall off the operator site. Lac repressor Lactose Lactose permease 2. lac operon proteins are synthesized. This promotes the efficient metabolism of lactose. 4. Most proteins involved with lactose utilization are degraded. Lac repressor 3. The lactose is depleted. Allolactose levels decrease. Allolactose is released from the repressor, allowing it to bind to the operator site. Figure 16.5a Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display Animation Lac Operon http://vcell.ndsu.nodak.edu/animations/ http://vcell.ndsu.nodak.edu/animations/ Experimental level Conceptual level Mutant + P O Z Y+ I– 1. Grow mutant strain and merozygote strain separately. Mutant strain Merozygote strain I– A+ + + PO Z Y + A 1. P I– + I+ P O ZY+ + F′ A O Z+ Y + A+ 2.Divide each strain into two tubes. Merozygote – Lactose Operon is constitutive-on in Mutant strain because no repressor is made. 2. P O Z +Y + A+ I– + Lactose – Lactose Lactose 3. In one of the two tubes, add lactose. 3. I– 1 2 3 + + PO Z Y + A 4 P O Z Y+ A+ F’ In mero-zygote strain, the lac I+ gene on the F´ factor makes enough repressor to bind to both operator sites (restoring WT phenotype on main chromosome). 4. Incubate the cells long enough to allow lac operon induction. 5. Burst the cells with a sonicator. This allows β-galactosidase to escape from the cells. + I+ 4. PO I– Z+Y+ A+ P I+ O Z+ Y+ + F A + Lactose Lactose is taken up, is converted to allolactose, and removes the repressor. Brooker Figure 16.7 6. Add β-o-nitrophenylgalactoside (β-ONPG). This is a colorless compound. β-galactosidase will cleave the compound to produce galactose and o-nitrophenol (O-NP). O-NP has a yellow color. The deeper the yellow color, the more β-galactosidase was produced. b-ONPG GalactoseO-NP b-o-nitrophenyl- 1. galactoside + NO2 NO2 Broken cell b-galactosidase + 2. NO2 3. 7. Incubate the sonicated cells to allow β-galactosidase time to cleave β-ONPG. NO2 1 2 3 4 4. + NO2 8. Measure the yellow color produced with a spectrophotometer. (See the Appendix for a description of spectrophotometry.) NO2 NO2 Brooker Figure 16.7, cont Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Table 16.1 16 – 34 Question Will a loss-of-function mutation in Plac (promoter sequence) be cis-dominant or trans-dominant? Lactose status Genotype Promoter Seq Repressor Operator Seq (assume absence of Glucose) Lac Z Lac Y Lac A Type of mutation (e.g. cisdominant, consititutive ON) Lac Z Absent WT + Active Bound No Expression No Expression No Expression none Present WT + Inactivated Open WT B-Gal WT Permease WT Transacet. none Present Lac Ymiss Present Lac ZNons Present P Lac(-) Absent Lac Oc Present Lac Oc Lactose status (assume absence of Glucose) Genotype Absent Lac I (-) Absent F’-Lac I (+) Lac I (-) Absent F’-LacOc Lac O+ Present F’-LacOc Lac O+ Promoter Seq Repressor Operator Seq Lac Z LacY Lac A Type of mutation (e.g. cisdominant) Go over lecture outline at end of lecture