* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download BIOINFORMATICS AND GENE DISCOVERY
Genomic library wikipedia , lookup
Transposable element wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Gene nomenclature wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene therapy wikipedia , lookup
Y chromosome wikipedia , lookup
Pathogenomics wikipedia , lookup
Minimal genome wikipedia , lookup
Gene desert wikipedia , lookup
Non-coding DNA wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genetic engineering wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Frameshift mutation wikipedia , lookup
Neocentromere wikipedia , lookup
Public health genomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Human genome wikipedia , lookup
Primary transcript wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics of human development wikipedia , lookup
History of genetic engineering wikipedia , lookup
X-inactivation wikipedia , lookup
Point mutation wikipedia , lookup
Genome editing wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene expression programming wikipedia , lookup
Designer baby wikipedia , lookup
Genome evolution wikipedia , lookup
Microevolution wikipedia , lookup
Genome (book) wikipedia , lookup
UNIVERSITY OF NORTH CAROLINA AT CHAPEL HILL
Bioinformatics Tutorials
BIOINFORMATICS
AND
GENE DISCOVERY
Iosif Vaisman
1998
From genes to proteins
From genes to proteins
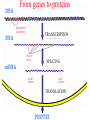
DNA
RNA
PROMOTER
ELEMENTS
TRANSCRIPTION
SPLICE
SITES
mRNA
SPLICING
START
CODON
STOP
CODON
TRANSLATION
PROTEI
From genes to proteins
Comparative Sequence Sizes
•
•
•
•
•
•
•
Yeast chromosome 3
Escherichia coli (bacterium) genome
Largest yeast chromosome now mapped
Entire yeast genome
Smallest human chromosome (Y)
Largest human chromosome (1)
Entire human genome
350,000
4,600,000
5,800,000
15,000,000
50,000,000
250,000,000
3,000,000,000
Low-resolution physical map
of chromosome 19
Chromosome 19 gene map
Computational Gene Prediction
•Where the genes are unlikely to be located?
•How do transcription factors know where to bind a region of DNA?
•Where are the transcription, splicing, and translation start and stop
signals?
•What does coding region do (and non-coding regions do not) ?
•Can we learn from examples?
•Does this sequence look familiar?
Artificial Intelligence in Biosciences
Neural Networks (NN)
Genetic Algorithms (GA)
Hidden Markov Models (HMM)
Stochastic context-free grammars (CFG)
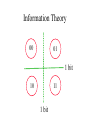
Information Theory
0
1
1 bit
Information Theory
00
01
1 bit
11
10
1 bit
Information Theory
1 bit
1 bit
Scientific Models
Physical models -- Mathematical models
Mechanistic models
Stochastic models
Mechanism
Black box
Predictive power
Elegance
Consistency
Predictive power
Hidden Markov models
Stochastic mechanism
Neural Networks
•interconnected assembly of simple processing elements (units or nodes)
•nodes functionality is similar to that of the animal neuron
•processing ability is stored in the inter-unit connection strengths (weights)
•weights are obtained by a process of adaptation to, or learning from, a set
of training patterns
Genetic Algorithms
Search or optimization methods using simulated evolution.
Population of potential solutions is subjected to
natural selection, crossover, and mutation
choose initial population
evaluate each individual's fitness
repeat
select individuals to reproduce
mate pairs at random
apply crossover operator
apply mutation operator
evaluate each individual's fitness
until terminating condition
Crossover
Parent A
Parent B
crossover point
Child AB
Child BA
Mutation
Markov Model (or Markov Chain)
A
T
C
T
A
G
Probability for each character based only on several
preceding characters in the sequence
# of preceding characters = order of the Markov Model
Probability of a sequence
P(s) = P[A] P[A,T] P[A,T,C] P[T,C,T] P[C,T,A] P[T,A,G]
Hidden Markov Models
States -- well defined conditions
Edges -- transitions between the states
T
G
A
A
C
C
T
ATGAC
ATTAC
ACGAC
ACTAC
Each transition asigned a probability.
Probability of the sequence:
single path with the highest probability --- Viterbi path
sum of the probabilities over all paths -- Baum-Welch method
Hidden Markov Model of Biased Coin Tosses
• States (Si):
Two Biased Coins {C1, C2}
• Outputs (Oj):
Two Possible Outputs {H, T}
• p(OutputsOij):
p(C1, H), p(C1, T), p(C2, H) p(C2, T)
• Transitions:
From State X to Y {A11, A22, A12, A21}
• p(Initial Si):
p(I, C1), p(I, C2)
• p(End Si):
p(C1, E), p(C2, E)
Hidden Markov Model for Exon and Stop Codon
(VEIL Algorithm)
GRAIL gene identification program
POSSIBLE EXONS
REFINED EXON
POSITIONS
FINAL EXON
CANDIDATES
Suboptimal Solutions for the Human Growth
Hormone Gene (GeneParser)
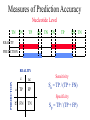
Measures of Prediction Accuracy
Nucleotide Level
TN
FN
TP
FP
TN
FN
TP
FN TN
REALITY
PREDICTION
c
c
TP
nc
FP
Sensitivity
Sn = TP / (TP + FN)
Specificity
nc
PREDICTION
REALITY
FN
TN
Sp = TP / (TP + FP)
Measures of Prediction Accuracy
Exon Level
MISSING
EXON
WRONG
EXON
CORRECT
EXON
Sn =
number of correct exons
number of actual exons
Sp =
number of correct exons
number of predicted exons
REALITY
PREDICTION
Sensitivity
Specificity
GeneMark Accuracy Evaluation
Bibliography
http://linkage.rockefeller.edu/wli/gene/list.html
and
http://www-hto.usc.edu/software/procrustes/fans_ref/
Gene Discovery Exercise
http://metalab.unc.edu/pharmacy/Bioinfo/Gene