* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download blumberg-lab.bio.uci.edu
Epigenetics of neurodegenerative diseases wikipedia , lookup
Non-coding DNA wikipedia , lookup
Transposable element wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene therapy wikipedia , lookup
Public health genomics wikipedia , lookup
Pathogenomics wikipedia , lookup
Oncogenomics wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression programming wikipedia , lookup
RNA silencing wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genomic imprinting wikipedia , lookup
Primary transcript wikipedia , lookup
Ridge (biology) wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genome evolution wikipedia , lookup
History of genetic engineering wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Microevolution wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome (book) wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Minimal genome wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Designer baby wikipedia , lookup
RNA interference wikipedia , lookup
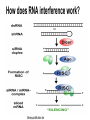
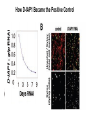
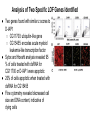
Timothy Abreo and Mrinal Sinha Goal: Ok, so we have the whole genome sequence of Drosophila Melanogaster and we have mapped its genes--now what? benchfly.com http://ocasapiens-dweb.blogautore.repubblica.it/ Model Organism--Drosophila Melanogaster (DM) We are going to look at very specific cell culture--particularly DM hemocytes Specific Mechanics RNAi = RNA Interference Specifically with dsRNA--now known as small interfering RNA (siRNA). quickmeme.com How does RNA interference work? Genequantification.de A specific example: Gene called D-IAP1. It is an anti-apoptosis gene. How D-IAP1 Became the Positive Control Functional Assays ● Flow Cytometry ● Sytox/Hoesht staining Ok, so what? galleryhip.com Well... We know what D-IAP1 does, and when we took it out, we saw our expected phenotype. But... We are going to look at very specific genes, specifically ones related to cellular growth and survival. scitechdaily.com Then we add this to a bunch of cells- Interpro Protein Domain Analysis Company’s Description: InterPro provides functional analysis of proteins by classifying them into families and predicting domains and important sites. We combine protein signatures from a number of member databases into a single searchable resource, capitalising on their individual strengths to produce a powerful integrated database and diagnostic tool. Phenotypic Screens ● 13 % Ribosomal genes and 8% Proteosome genes found ● Cell cycle genes only found significant in S2-R cells ● Most populated section with 14% were genes encoding proteins with DNA binding domains ● DNA binding domains included chromatin related factors and transcription factors Analysis of Essential Genes Functions ● Focused on 438 transcripts with z scores >3 ○ only 20% of these genes had associated mutant alleles ○ 47% had associated gene ontologies ○ Interpro: 59% identified protein domains, 37% in severe cases Analysis of Two Specific LOF Genes Identified ● Two genes found with similar z scores to D-IAP1 ○ CG11700: ubiquitin-like gene ○ CG15455: encodes acute myeloid leukemia-like transcription factor ● Sytox and Hoesht analysis revealed 95 % of cells treated with dsRNA for CG11700 or D-IAP1 were apoptotic ● 20% of cells apoptotic when treated with dsRNA for CG15455 ● Flow cytometry revealed decreased cell size and DNA content, indicative of dying cells Rescue of the Two LOF Phenotypes ● Cells treated with an inhibitor of proapoptotic caspase (Nedd2-like) reverted the cell death in response to RNAi for either CG11700, DIAP1, or (to a lesser extent) CG15455 ● Same phenomena was observed when cells were treated with RNAi targeted at proapoptotic caspases, except in cells with coRNAi for CG15455 ● Demonstrated to researchers that CG11700 and D-IAP1 were likely involved in the same pathway Ortholog and Homolog Search ● Percentage of predicted orthologs with RNAi analysis was higher than those found by database searches ● 50 genes had homology to human disease genes, with 10 associated with blood-cell leukemia o Demonstrated that functional RNAi analysis in Drosophila can discover common genes involved in cell viability of Animals ● However, in severe RNAi phenotypes, few homologs were found identified amongst yeast and animals o Suggests that metazoans evolved specific mechanisms involved in cell viability Implications and Future Applications ● Functional RNAi analysis can reveal unknown gene and evolutionary conserved gene functions ● Quantitative nature of this analysis facilitates detection of gene functions with subtle or redundant phenotypes via inducing a LOF ● Allows for statistical clustering in comparing several cellular phenotypes to determine gene functions ● Genome wide RNAi can be utilized to screen multiple cellular pathways and their various components, allowing for a better understanding of systems involved in disease and development Thanks...for staying awake! Any Questions?