* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Introduction to bioinformatics
Gene desert wikipedia , lookup
Non-coding RNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Transposable element wikipedia , lookup
History of RNA biology wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene nomenclature wikipedia , lookup
Protein moonlighting wikipedia , lookup
Public health genomics wikipedia , lookup
Designer baby wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Microevolution wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genomic library wikipedia , lookup
Non-coding DNA wikipedia , lookup
Human Genome Project wikipedia , lookup
Human genome wikipedia , lookup
Multiple sequence alignment wikipedia , lookup
Point mutation wikipedia , lookup
Pathogenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome evolution wikipedia , lookup
Sequence alignment wikipedia , lookup
Genome editing wikipedia , lookup
Helitron (biology) wikipedia , lookup
Metagenomics wikipedia , lookup
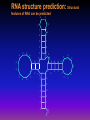
BioInformatics - What and Why? The following power point presentation is designed to give some background information on Bioinformatics. This presentation is modified from information supplied by Dr. Bruno Gaeta, and with permission from eBioInformatics Pty Ltd (c) Copywright The need for bioinformaticists. The number of entries in data bases of gene sequences is increasing exponentially. Bioinformaticians are needed to understand and use this information. GenBank growth 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 Genome sequencing projects, including the human genome project are producing vast amounts of information. The challenge is to use this information in a useful way Publically available genomes (April 1998) COMPLETE/PUBLIC Aquifex aeolicus Pyrococcus horikoshii Bacillus subtilis Treponema pallidum Borrelia burgdorferi Helicobacter pylori Archaeoglobus fulgidus Methanobacterium thermo. Escherichia coli Mycoplasma pneumoniae Synechocystis sp. PCC6803 Methanococcus jannaschii Saccharomyces cerevisiae Mycoplasma genitalium Haemophilus influenzae COMPLETE/PENDING PUBLICATION Rickettsia prowazekii Pseudomonas aeruginosa Pyrococcus abyssii Bacillus sp. C-125 Ureaplasma urealyticum Pyrobaculum aerophilum ALMOST/PUBLIC Pyrococcus furiosus Mycobacterium tuberculosis H37Rv Mycobacterium tuberculosis CSU93 Neisseria gonorrhea Neisseria meningiditis Streptococcus pyogenes Terry Gaasterland, Siv Andersson, Christoph Sensen http://www.mcs.anl.gov/home/gaasterl/genomes.html Bioinformatics impacts on all aspects of biological research. ”..We must hook our individual computers into the worldwide network that gives us access to daily changes in the databases and also makes immediate our communications with each other. The programs that display and analyze the material for us must be improved - and we must learn to use them more effectively. Like the purchased kits, they will make our life easier, but also like the kits, we must understand enough of how they work to use them effectively…” Walter Gilbert (1991) “Towards a paradigm shift in biology” Nature News and Views 349:99 Promises of genomics and bioinformatics Medicine Knowledge of protein structure facilitates drug design Understanding of genomic variation allows the tailoring of medical treatment to the individual’s genetic make-up Genome analysis allows the targeting of genetic diseases The effect of a disease or of a therapeutic on RNA and protein levels can be elucidated The same techniques can be applied to biotechnology, crop and livestock improvement, etc... What is bioinformatics? Application of information technology to the storage, management and analysis of biological information Facilitated by the use of computers What is bioinformatics? Sequence analysis Molecular modeling Geneticists obtain information about the evolution of organisms by looking for similarities in gene sequences Ecology and population studies Crystallographers/ biochemists design drugs using computer-aided tools Phylogeny/evolution Geneticists/ molecular biologists analyse genome sequence information to understand disease processes Bioinformatics is used to handle large amounts of data obtained in population studies Medical informatics Personalised medicine Sequence analysis: overview Sequencing project management Nucleotide sequence analysis Sequence entry Sequence database browsing Manual sequence entry Nucleotide sequence file Search for protein coding regions Search databases for similar sequences Design further experiments Restriction mapping PCR planning coding non-coding Protein sequence analysis Translate into protein Search databases for similar sequences Sequence comparison Search for known motifs RNA structure prediction Create a multiple sequence alignment Edit the alignment Molecular phylogeny Search for known motifs Predict secondary structure Sequence comparison Multiple sequence analysis Format the alignment for publication Protein sequence file Protein family analysis Predict tertiary structure Gene Sequencing: Automated chemcial sequencing methods allow rapid generation of large data banks of gene sequences Database similarity searching: The BLAST program has been written to allow rapid comparison of a new gene sequence with the 100s of 1000s of gene sequences in data bases Sequences producing significant alignments: (bits) Value gnl|PID|e252316 (Z74911) ORF YOR003w [Saccharomyces cerevisiae] 112 gi|603258 (U18795) Prb1p: vacuolar protease B [Saccharomyces ce... 106 gnl|PID|e264388 (X59720) YCR045c, len:491 [Saccharomyces cerevi... 69 gnl|PID|e239708 (Z71514) ORF YNL238w [Saccharomyces cerevisiae] 30 gnl|PID|e239572 (Z71603) ORF YNL327w [Saccharomyces cerevisiae] 29 gnl|PID|e239737 (Z71554) ORF YNL278w [Saccharomyces cerevisiae] 29 7e-26 5e-24 7e-13 0.66 1.1 1.5 gnl|PID|e252316 (Z74911) ORF YOR003w [Saccharomyces cerevisiae] Length = 478 Score = 112 bits (278), Expect = 7e-26 Identities = 85/259 (32%), Positives = 117/259 (44%), Gaps = 32/259 (12%) Query: 2 QSVPWGISRVQAPAAHNRG---------LTGSGVKVAVLDTGIST-HPDLNIRGG-ASFV 50 + PWG+ RV G G GV VLDTGI T H D R + + Sbjct: 174 EEAPWGLHRVSHREKPKYGQDLEYLYEDAAGKGVTSYVLDTGIDTEHEDFEGRAEWGAVI 233 Query: 51 PGEPSTQDGNGHGTHVAGTIAALNNSIGVLGVAPSAELYXXXXXXXXXXXXXXXXXQGLE 110 P D NGHGTH AG I + + GVA + ++ +G+E Sbjct: 234 PANDEASDLNGHGTHCAGIIGSKH-----FGVAKNTKIVAVKVLRSNGEGTVSDVIKGIE 288 Sequence comparison: Gene sequences can be aligned to see similarities between gene from different sources 768 TT....TGTGTGCATTTAAGGGTGATAGTGTATTTGCTCTTTAAGAGCTG || || || | | ||| | |||| ||||| ||| ||| 87 TTGACAGGTACCCAACTGTGTGTGCTGATGTA.TTGCTGGCCAAGGACTG . . . . . 814 AGTGTTTGAGCCTCTGTTTGTGTGTAATTGAGTGTGCATGTGTGGGAGTG | | | | |||||| | |||| | || | | 136 AAGGATC.............TCAGTAATTAATCATGCACCTATGTGGCGG . . . . . 864 AAATTGTGGAATGTGTATGCTCATAGCACTGAGTGAAAATAAAAGATTGT ||| | ||| || || ||| | ||||||||| || |||||| | 173 AAA.TATGGGATATGCATGTCGA...CACTGAGTG..AAGGCAAGATTAT 813 135 863 172 913 216 Restriction mapping: Genes can be analysed to detect gene sequences that can be cleaved with restriction enzymes 50 AceIII AluI AlwI ApoI BanII BfaI BfiI BsaXI BsgI BsiHKAI Bsp1286I BsrI BsrFI CjeI CviJI CviRI DdeI DpnI EcoRI HinfI MaeIII MnlI MseI MspI NdeI Sau3AI SstI TfiI Tsp45I Tsp509I TspRI 100 150 200 250 1 2 1 2 1 2 1 1 1 1 1 2 1 2 4 1 2 2 1 2 1 1 2 1 1 2 1 2 1 3 1 CAGCTCnnnnnnn’nnn... AG’CT GGATCnnnn’n_ r’AATT_y G_rGCy’C C’TA_G ACTGGG ACnnnnnCTCC GTGCAGnnnnnnnnnnn... G_wGCw’C G_dGCh’C ACTG_Gn’ r’CCGG_y CCAnnnnnnGTnnnnnn... rG’Cy TG’CA C’TnA_G GA’TC G’AATT_C G’AnT_C ’GTnAC_ CCTCnnnnnn_n’ T’TA_A C’CG_G CA’TA_TG ’GATC_ G_AGCT’C G’AwT_C ’GTsAC_ ’AATT_ CAGTGnn’ PCR Primer Design: Oligonucleotides for use in the polymerisation chain reaction can be designed using computer based prgrams OPTIMAL primer length MINIMUM primer length MAXIMUM primer length OPTIMAL primer melting temperature MINIMUM acceptable melting temp MAXIMUM acceptable melting temp MINIMUM acceptable primer GC% MAXIMUM acceptable primer GC% Salt concentration (mM) DNA concentration (nM) MAX no. unknown bases (Ns) allowed MAX acceptable self-complementarity MAXIMUM 3' end self-complementarity GC clamp how many 3' bases --> --> --> --> --> --> --> --> --> --> --> --> --> --> 20 18 22 60.000 57.000 63.000 20.000 80.000 50.000 50.000 0 12 8 0 Gene discovery: Computer program can be used to recognise the protein coding regions in DNA 0 1,000 2,000 3,000 4,000 0 1,000 2,000 3,000 4,000 2.0 1.5 1.0 0.5 -0.0 2.0 1.5 1.0 0.5 -0.0 2.0 1.5 1.0 0.5 -0.0 Plot created using codon preference (GCG) RNA structure prediction: Structural features of RNA can be predicted A C G U A G A U G C U A C A U A C A C G G GU C G U GA A U U C U A G U G C G G G U A A C C G UC G U C C A G G U A G U G CG A U C C U G C G C C A C Protein structure prediction: Particular structural features can be recognised in protein sequences 50 100 50 100 5.0 KD Hydrophobicity -5.0 10 Surface Prob. 0.0 1.2 Flexibility 0.8 1.7 Antigenic Index -1.7 CF Turns CF Alpha Helices CF Beta Sheets GOR Turns GOR Alpha Helices GOR Beta Sheets Glycosylation Sites Protein Structure : the 3-D structure of proteins is used to understand protein function and design new drugs Multiple sequence alignment: Sequences of proteins from different organisms can be aligned to see similarities and differences Alignment formatted using MacBoxshade Phylogeny inference: Analysis of sequences allows evolutionary relationships to be determined E.coli C.botulinum C.cadavers C.butyricum B.subtilis B.cereus Phylogenetic tree constructed using the Phylip package Large scale bioinformatics: genome projects Mapping Identifying the location of clones and markers on the chromosome by genetic linkage analysis and physical mapping Using database searches, pattern searches, protein family analysis and structure prediction to assign a function to each predicted gene Data mining Searching for relationships and correlations in the information Sequencing Assembling clone sequence reads into large (eventually complete) genome sequences Gene discovery Identifying coding regions in genomic DNA by database searching and other methods Function assignment Genome comparison Comparing different complete genomes to infer evolutionary history and genome rearrangements Challenges in bioinformatics Explosion of information Need for faster, automated analysis to process large amounts of data Need for integration between different types of information (sequences, literature, annotations, protein levels, RNA levels etc…) Need for “smarter” software to identify interesting relationships in very large data sets Lack of “bioinformaticians” Software needs to be easier to access, use and understand Biologists need to learn about the software, its limitations, and how to interpret its results