* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Intensity-Dependent Normalization
Genetically modified crops wikipedia , lookup
Genetics and archaeogenetics of South Asia wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genome evolution wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genetic testing wikipedia , lookup
Gene expression programming wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Non-coding DNA wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
X-inactivation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genetic engineering wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Genomic imprinting wikipedia , lookup
Public health genomics wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Human genetic variation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Genetic drift wikipedia , lookup
Behavioural genetics wikipedia , lookup
Medical genetics wikipedia , lookup
Designer baby wikipedia , lookup
Heritability of IQ wikipedia , lookup
Population genetics wikipedia , lookup
Microevolution wikipedia , lookup
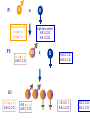
Course on Biostatistics • • • • Instructors – Dr. Małgorzata Bogdan Dr. David Ramsey Institute of Mathematics and Computer Science • Wrocław University of Technology • Poland Course on Biostatistics • • • • • Two parts 1. Locating genes influencing quantitative traits in experimental populations. 20.03-30.03. 2006, Małgorzata Bogdan 2. Population Genetics 22.05-2.06.2006, David Ramsey Grading • Students can gain 50 points for each part of the course (25 for a project, 25 for an exam). • The final grade will be based on the total percentage. • To pass the course the student has to gain at least 15 points for each part of the course and at least 50 points in total. First part • Locating genes influencing quantitative traits in experimental populations. • 20.03-30.03. 2006 Małgorzata Bogdan Course Outline • • • Introduction to genetics and experimental populations. Basic methods of locating quantitative trait loci (QTL). Locating QTL with QTL Cartographer. Helpful materials • Text book • Genetics and Analysis of Quantitative Traits by Michael Lynch and Bruce Walsh • Software – Windows QTL Cartographer • S.Wang, C.J. Basten, Z-B. Zeng • Program in Statistical Genetics, North Carolina State University • Can be downloaded from • http://statgen.ncsu.edu/qtlcart/WQTLCart.htm Main Goal • Learn how to locate regions of the genome hosting genes influencing some quantitative traits (Quantitative Trait Loci – QTL). • Statistical methods – mainly linear models. Introduction to Genetics DNA - A nucleic acid that carries the genetic information in the cell. DNA consists of two long chains of nucleotides joined by hydrogen bonds between the complementary bases adenine and thymine or cytosine and guanine. The sequence of nucleotides determines individual hereditary characteristics. http://www.answers.com/topic/dna Introduction to Genetics • Chromosome – a ‘’long’’, continuous piece of DNA, which contains many genes, regulatory elements and other intervening nucleotide sequences. • Diploid organisms – chromosomes appear in pairs (one from each parent) • Allele - any of two or more alternative forms of a gene that occupy the same locus on a chromosome. • Example: allele of blue eyes, allele of brown eyes • Genotype at a single locus: the pair of alleles that individual carries at the locus. The Hardy-Weinberg principle • • • • • • Random mating pa- frequency of a allele pA- frequency of A allele P(aa)=p2a P(aA)=2papA P(AA)=p2A • Phenotype – observed or measured characteristic (or trait) for an individual. • We will be dealing with quantitative traits like eg. height, yield, blood pressure etc. Heritability • Z - the phenotypic (trait) value of an individual • G – the genotypic value (the sum of the total effects of all loci on the trait) • E – an environmental deviation • Z=G+E • Broad sense heritability (population parameter) • H2 = Var (G) / Var (Z) The influence of a single locus Fisher’s decomposition of the Genotypic Value • Consider a biallelic locus with alleles a, A • N – number of alleles ’’a’’ for a given individual (gene content) • We regress G on N G N Gˆ - the average effect of allelic substituti on •Var(G) = 2A+ 2D Trait Influenced by Two Loci • Gijkl – mean phenotype for individuals with genotypes (i j; k l) • αi = Gi…- G - additive effect of i allele • δij = Gij..- G - αi - αj - dominance effects at the first locus • δkl = Gkl..- G - αk - αl - dominance effects at the second locus Possible interactions (epistasis) • (αα)ik=Gi.k.- G – αi – αk • (αδ)ikl= • Gi.kl.- G – αi – αk – αl – δkl- (αα)ik- (αα)il • (δδ)ijkl= Gijkl.- G – αi – αj – αk – αl – δij - δkl - (αα)ik- (αα)il-(αα)jk- (αα)jl - (αδ)ikl- (αδ)jkl - (αδ)ijk- (αδ)ijl Example (Lynch and Walsh) • Teosinte – wild progenitor of cultivated maize • Two loci (markers) - UMC107 , BV302 • UM, BM – maize alleles • UT, BT – teosinte alleles • Trait – the average length of the vegetative internodes in the lateral branch (in mm) Mean trait values UMC107 BV302 UMM UMT UTT BMM 18.0 40.9 61.1 BMT 54.6 47.6 66.5 BTT 47.8 83.6 101.7 Genotype Frequencies UMC107 BV302 BMM BMT BTT UMM UMT UTT Cockerham model 1 1 if X 0 if 1 if AA aA aa 1 / 2 if AA or aa Z 1/2 if Aa G X Z Cockerham model 2 GAA 1 - 1 - 1/2 G 1 0 1/2 aA Gaa 1 1 - 1/2 Cockerham model 3 two loci Gij 1 X 1i 2 X 2i 1Z1i 2 Z 2i aa X 1i X 2i ad X 1i Z 2i da Z1i X 2i dd Z1i Z 2i Genetic maps Markers – genetic loci which express experimentally detectable variation between individuals. Genetic map gives an order of markers on a chromosome and a distance between them. 1 Morgan – the expected value of the number of crossovers is equal to 1 Genetic map A P: x low fat content abqcd abqcd F1: a b q c d ABQCD B high fat content ABQCD ABQCD AB x B ABQCD ABQCD BC: a b q c d ABQCD AB q c d ABQCD a BQC d ABQCD ABQCD ABQCD