* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Class I tRNA
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Drug design wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Point mutation wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Western blot wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Ligand binding assay wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Polyadenylation wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Homology modeling wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Metalloprotein wikipedia , lookup
Proteolysis wikipedia , lookup
Protein structure prediction wikipedia , lookup
Gene expression wikipedia , lookup
Messenger RNA wikipedia , lookup
Community fingerprinting wikipedia , lookup
Genetic code wikipedia , lookup
Biosynthesis wikipedia , lookup
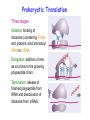
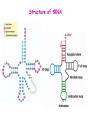
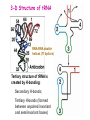
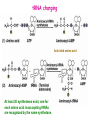
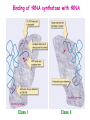
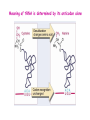
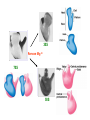
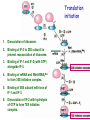
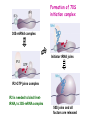
Prokaryotic Translation Three stages Initiation: binding of ribosome (containing rRNAs and proteins) and aminoacyl tRNA to mRNA. Elongation: addition of one aa at a time to the growing polypeptide chain. Termination: release of finished polypeptide from tRNA and dissociation of ribosome from mRNA. Preinitiation: tRNA charging; Dissociation of ribosome Structure of tRNA D Structure of aminoacyl-tRNA Length: 76 (74-95) residues. Extra arm (variable loop): Class I tRNA: 3-5 bases Class II tRNA: 13-21 bases; ~5 bases in the stem. Additional types of base pairing: G·U, G·Y, A·Y. Less stable Positions: Invariant: maintain 2O structure Semiinvariant (other Pu or Py) Name of tRNA: Tyr tRNA1 Tyr tRNA2 Name of charged tRNA: Tyr-tRNA A C C I tRNA are processed from longer precursors Modified nucleosides found in tRNA The bases are modified (>50 different types) after transcription by specific tRNAmodifying enzymes to affect the efficiency of charging and pairing properties. Y 3-D Structure of tRNA RNA-RNA double helices (11 bp/turn) Tertiary structure of tRNA is created by H-bonding: Secondary H-bonds; Tertiary H-bonds (formed between unpaired invariant and semiinvariant bases) tRNA charging Activated amino acid At least 20 synthetases exist, one for each amino acid. Isoaccepting tRNAs are recognized by the same synthetase. tRNA charging Recognition depends on an interaction between a few points of contact in tRNA, mostly at the acceptor stem and anticodon, and a few amino acids constituting the active site in the enzyme. Binding of tRNA synthetase with tRNA Two classes of tRNA synthetase: Class I tRNA synthetases Aminoacylate the 2’-OH group of the terminal A of the tRNA. Approach the tRNA from the D-loop and acceptor stem minor groove side. Class II tRNA synthetases Aminoacylate the 3’-OH group of the terminal A of the tRNA. Approach the tRNA from the variable arm and acceptor stem major groove side (the opposite side of tRNA that contacts the class I enzyme). tRNA Synthetase Location of varies Each class contains about 10 enzymes Binding of tRNA synthetase with tRNA Class I Class II Recognition of correct tRNA by tRNA synthetase is achieved by two steps: Association Aminoacylation Recognition of correct amino acid by tRNA synthetase is also achieved by two steps (every synthetase undergoes proofreading at either stage), which occurs only in the presence of cognate tRNA. Ile-tRNA synthetase has two active sites for sieving the cognate amino acid Synthetic site: activation of amino acid Editing site: hydrolysis of incorrrect aminoacyl-tRNA Accuracy of charging tRNAIle by its synthetase depends on error control by two steps Meaning of tRNA is determined by its anticodon alone Structure of Ribosome r- 30S Remove Mg 2+ 70S 50S Arrangement of proteins and 16S rRNA in S30 subunit Central domain RNA is concentrated at the interface with the 50S subunit. Both 30S and 50S subunits are self-assembled in vitro. In 30S subunit, S4 and S8 bind to 16S rRNA first, other proteins then join sequentially and cooperatively. 16S rRNA Secondary structure of 16S rRNA and interaction of this RNA with proteins and tRNA were studied by primer extension and crosslinking, and other techniques. Features of rRNAs rRNAs have considerable 2o structure (This is analyzed by comparing the sequences of corresponding rRNAs in related organisms). About 2% of the residues in rRNAs are methylated, which may be important for ribosomal function. Interaction of rRNA with some ribosomal protein induce conformational change of rRNA so that it can interact with another protein. rRNA interacts with mRNA or tRNA at each stage of translation. The proteins are necessary only to maintain the rRNA in a structure in which rRNA can perform the catalytic function. Conformation of rRNAs is flexible during protein synthesis. The 3’ terminus of 16S rRNA pairs with the SD sequence of mRNA at initiation. rRNA contacts the tRNA at parts of the structure that are universally conserved. Translation initiation 1. Dissociation of ribosome. 2. Binding of IF-3 to 30S subunit to prevent reassociation of ribosome. 3. Binding of IF-1 and IF-2 (with GTP) alongside IF-3. 4. Binding of mRNA and fMet-tRNAfMet to form 30S initiation complex. 5. Binding of 50S subunit with loss of IF-1 and IF-3. 6. Dissociation of IF-2 with hydrolysis of GTP to form 70S initiation complex. Recycle of ribosome and initiation factors Ribosomes bind to mRNA at a special sequence Ribosomal protein S12 and 16S rRNA, are responsible for recognition of the SD sequence. A conserved sequence near the 3’-end of 16S rRNA pairs with the SD sequence. mRNA sequence protected by ribosome (about 30 nt) Ribosomal binding to the SD sequence provides a means for controlling gene expression. IF-3 is the primary factor for mRNA binding to ribosome. IF-1 and IF-2, which bind near IF-3, assist assembly of 30S initiation complex. fMet-tRNAf as initiator tRNA This initiator tRNA recognizes codons AUG (90%), GUG (8%), or UUG (1%) that lies within a ribosome-binding site. Met tRNAf is different Met from tRNAm ; the former goes to the first AUG codon and the methionine of the later can not be formylated. Formation of fMet-tRNAf Formylation is not strictly necessary for the initiator tRNA to function in initiation. It is the tRNA part of fMet-tRNAf that makes it the initiating aminoacyltRNA. IF-2 ensures only the initiator tRNA goes to the P-site at initiation. Features of Met fMet-tRNAf Formation of 70S initiation complex 30S-mRNA complex Initiator tRNA joins IF2-GTP joins complex IF2 is needed to bind fmettRNAf to 30S-mRNA complex 50S joins and all factors are released Removal of formyl group Deformylase Removal of methionine Aminopeptidase In bacteria and mitochondria, formyl group, and sometimes the methionine, is removed during translation.