* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Review 3
Epitranscriptome wikipedia , lookup
Butyric acid wikipedia , lookup
Nitrogen cycle wikipedia , lookup
Plant nutrition wikipedia , lookup
Peptide synthesis wikipedia , lookup
Biochemical cascade wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Phosphorylation wikipedia , lookup
Electron transport chain wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Metalloprotein wikipedia , lookup
Microbial metabolism wikipedia , lookup
Genetic code wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Photosynthesis wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Biosynthesis wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Biochemistry wikipedia , lookup
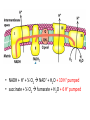
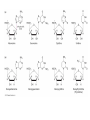
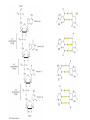
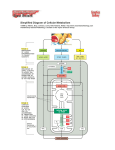
Review 3 C483 Spring 2013 Net outcomes of • • • • • Oxidative phosphorylation (shuttles) Fatty acid catabolism Nitrogen catabolism Light reactions of photosynthesis Calvin cycle Structures to know • All amino acids • (deoxy)ribonucleosides and (deoxy)ribonucleotides • Carbamoyl phosphate and urea • Pyruvate, oxaloacetate, a-ketoglutarate • PRPP Metabolic Pathways/Reactions • Know these reactions/pathways (not arrow mechanisms) – Beta oxidation – PLP-mediated transamination – Formation of carbamoyl phosphate – Catabolism of amino acids covered in lecture – Basic hydrolysis and RNase-catalyzed hydrolysis of RNA Protonmotive Force • NADH + H+ + ½ O2 NAD+ + H2O + 10 H+ pumped • succinate + ½ O2 fumarate + H2O + 6 H+ pumped Photosynthesis: Net Reaction 2 H2O + 4 NADP+ + 8 photons O2 + 4 NADPH Proton Gradient • For every ONE O2 – 4 e- passed – 8 photons absorb – 12 H+ pumped • In plants, 12 H+ required to turn ATP synthase – 3 ATP made • Quantum yield is 3 ATP/8 photons or ATP/2.7 photon