* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download PowerPoint-presentatie
Phosphorylation wikipedia , lookup
Signal transduction wikipedia , lookup
List of types of proteins wikipedia , lookup
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein domain wikipedia , lookup
Homology modeling wikipedia , lookup
Cooperative binding wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Western blot wikipedia , lookup
Protein purification wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
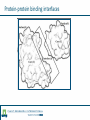
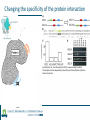
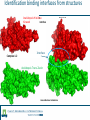
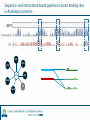
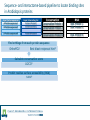
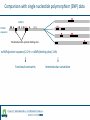
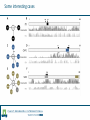
Identification of protein-protein binding motifs Felipe Leal Valentim Aalt-Jan van Dijk [email protected] [email protected] Plant Research International Applied Bioinformatics Protein-protein binding interfaces Protein-protein binding interfaces Surface Surface Interface Ligand binding site Core Core Core structural residues Properties: DNA-binding site Exposed in the protein surface; Functionally/Structurally important residues are more highly conserved; Changing the specificity of the protein interaction [van Dijk AD et al., PLoS Comput Biol. 2010] - Sequence Motifs in MADS Transcription Factors Responsible for Specificity and Diversification of ProteinProtein Interaction Protein-protein binding motifs Interface Protein-protein binding motifs Protein binding interfaces are composed by residues highly conserved and exposed in the surface; The interface can be represented by short sequence motifs; which are thought to be overrepresented in pairs of interacting proteins. Identification binding interfaces from structures Protein 1 Arabidopsis Protein Histidine 1 Binding interface Protein 2 Kinase4 Interface Complex 1-2 Arabidopsis Trans Zeatin [Hubbard SJ, Thornton JM] Naccess V2.1.1 - Atomic Solvent Accessible Area Calculations Protein 2 Binding interface Structural information available in the PDB Sequence- and interactome-based pipeline to locate binding sites in Arabidopsis proteins Sequences -> The evolutionary conservation; Sequences -> Residue surface accessibility; Interactome -> Overrepresented motifs; Motif that are: likely to be exposed in the surface; conserved across species; and overrepresented in pairs of interacting proteins. Sequence- and interactome-based pipeline to locate binding sites in Arabidopsis proteins IAA7 IAA2 IAA11 SHY2 IAA1 IAA16 TPL IAA18 Sequence- and interactome-based pipeline to locate binding sites in Arabidopsis proteins Protein1-Protein2 >Protein sequence2 Protein2-Protein4 ... >Protein sequenceN ProteinN-ProteinM Conservation Protein N Find orthlogs from each protein sequence OrthoMCL1 Best blast reciprocal hint2 Calculate conservation score Al2CO3 Predict residue surface accessibility (RSA) SABLE4 RSA RSA Protein 1 RSA Protein 2 .. . >Protein sequence1 Conservation Conservation Protein 1 Conservation Protein 2 .. . Input Interacting list ... Input fasta sequences RSA Protein N Assessment of the pipeline's performance Non-interface motifs Predicted motifs False Positives (FP) Precision = TP/(TP + FP) Interface motif True Positives (TP) Assessment of the pipeline's performance Coverage: up to 42%, 22% and 42%, respectively for the human, yeast and Arabidopsis subsets. Precision: up to 58%, 96% and 100%. Locating interaction binding sites in Arabidopsis sequences at a large scale – Overview Predicted motifs: 1498 interactions among 985 proteins 36% of the proteins in the interactome and ~5.5% of all Arabidopsis proteins Validation and bioinformatics analysis Comparison with single nucleotide polymorphism (SNP) data nsSNP’s Protein sequence Predicted protein-protein binding sites nsSNPs(protein sequence):2.2% > nsSNPs(binding sites):1.6% Functional constraints Intermolecular coevolution Comparison with annotation of amino acid mutagenesis amino acid mutagenesis Proteins with a predicted motif n=985 Protein sequence Others functionally important sites Protein-protein binding sites DNA binding sites Mutagenesis annotation (UniProt) (n=38) 16 cases: predicted motifs overlap the mutated amino acid Some interesting cases Master's Project Proposal: Cross-species analysis of proteinprotein binding motifs Question??????? Practical assignment – Perl scripting for