* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download 3D Structure - Canadian Bioinformatics Workshops
Artificial gene synthesis wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
Biochemical cascade wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Interactome wikipedia , lookup
Paracrine signalling wikipedia , lookup
Magnesium transporter wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Signal transduction wikipedia , lookup
Protein purification wikipedia , lookup
Gene expression wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Western blot wikipedia , lookup
Protein structure prediction wikipedia , lookup
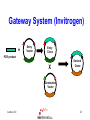
Protein–protein interaction wikipedia , lookup
Proteolysis wikipedia , lookup
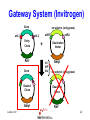
Protein Expression, Structural Proteomics & Bioinformatics David Wishart University of Alberta Edmonton, AB [email protected] Lecture 3.0 1 Expression Questions • • • • • • • • Which host cell system? Which expression vector? Which cloning/expression protocols? Is it membrane or water soluble? Is it single domain or multi-domain? How soluble and how stable? Where will this protein be found? How to purify & how to identify? Lecture 3.0 2 Host Cell System? • • • • • • • • • Escherichia coli Other bacteria Pichia pastoris Other yeast Baculovirus Animal cell culture Plants Sheep/cows/humans Cell free Lecture 3.0 Polyhedra 3 Host Cell System? • Choice depends on size and character of protein – Large proteins (>100 kD)? Choose eukaryote – Small proteins (<30 kD)? Choose prokaryote – Glycosylation essential? Choose baculovirus or mammalian cell culture – Isotopic labelling esential? Choose E. coli – Post-translational modifications essential? Choose yeast, baculovirus or other eukaryote Lecture 3.0 4 Host Cell System? • Try different hosts when optimizing expression (protease negative, strains with enhanced expression of rare tRNAs) • Expression levels can vary by a factor of 10 or more depending on strain choice • Example E. coli strains – MC1061, UT580, GM48, JM101, DH5, MG1065, NM522, MC4100, TOP10F’, BL21(DE3) BL21CodonPlus (DE3) Lecture 3.0 5 Codon Bias http://www.kazusa.or.jp/codon/ Lecture 3.0 6 Arginine Codon Bias E. coli AGA 2.7 AGG 1.6 Eubacteria (rare) Lecture 3.0 M. jannaschii AGA 27.5 AGG 9.9 Archaebacteria (abundant) H. sapiens AGA 11.2 AGG 11.1 Eukaryote (normal) 7 Host Cell System? • American Type Culture Collection – http://www.atcc.org • Clontech Cell Lines – http://www.clontech.com • Stratagene Cells (BL21) – http://stratagene.com • Invitrogen Cell Lines (Pichia) – http://www.invitrogen.com Lecture 3.0 8 Fermentor or Shake Flask? Lecture 3.0 9 Media Optimization • Still using L-broth? Try using T-broth – Tryptone - 12 g, Yeast Extract - 24 g, glycerol - 4 ml, KH2PO4 - 2.3g, K2HPO4 - 12.5g • Extra Spicy Media – More ATP: 10 ml/L glycerol + 10g glucose/L – More AA: Add 10g casamino acids + 10mg L-Trp • Add more media (30%) when you induce • Add more antibiotic when you induce – prevents overgrowth by cells that lost plasmid Lecture 3.0 10 Expression Questions • • • • • • • • Which host cell system? Which expression vector? Which cloning/expression protocols? Is it membrane or water soluble? Is it single domain or multi-domain? How soluble and how stable? Where will this protein be found? How to purify & how to identify? Lecture 3.0 11 Which Vector? • Must be compatible with host cell system (prokaryotic vectors for prokaryotic cells, eukaryotic vectors for eukaryotic cells) • Needs a good combination of – – – – – strong promoters ribosome binding sites termination sequences affinity tag or solubilization sequences multi-enzyme restriction site Lecture 3.0 12 Which Vector? • Promoters – arabinose systems (pBAD), phage T7 (pET), Trc/Tac promoters, phage lambda PL or PR • Tags – – – – – – – His6 for metal affinity chromatography (Ni) FLAG epitope tage DYKDDDDK CBP-calmodulin binding peptide (26 residues) E-coil/K-coil tags (poly E35 or poly K35) c-myc epitope tag EQKLISEEDL Glutathione-S-transferase (GST) tags Cellulose binding domain (CBD) tags Lecture 3.0 13 Which Vector? • VectorDB – http://vectordb.atcg.com • Invitrogen Vectors – http://www.invitrogen.com/vectors.html • Qiagen Vectors – http://www.qiagen.com/literature/vectors.asp • Stratagene Vectors – http://stratagene.com/vectors/vectors.htm Lecture 3.0 14 Cloning Software • • • • • • • MacVector (Accelrys) SimVector (Premier BioSoft) GeneTool (BioTools) Vector NTI (Informax/Invitrogen) DNAStrider LaserGene (DNAStar) PlasMapper (Bioinformatics Help Desk) Lecture 3.0 15 PlasMapper http://wishart.biology.ualberta.ca/PlasMapper/ Lecture 3.0 16 How to Clone? Echo Cloning Lecture 3.0 17 How to Clone? Yeast Cells Lecture 3.0 18 How to Clone? Mammalian Cells Lecture 3.0 19 Gateway System (Invitrogen) • No need to design, construct or ID unique restriction sites • Uses lambda phage site-specific recombination for gene/plasmid integration • No need for restriction enzyme digestions • No need for gel fragment separation and purification • Ideal for high throughput proteomics efforts Lecture 3.0 20 Gateway System (Invitrogen) + Entry Vector Entry Clone PCR product X Desired Clone Destination Vector Lecture 3.0 21 Gateway System (Invitrogen) Gene attR1 attL2 attL1 Entry Clone Kmr Gene Desired Clone Ampr Lecture 3.0 -ve selector (anti-gyrase) attR2 Destination Vector + Int IHF Xis Ampr -ve selector (anti-gyrase) Dead-end Clone Kmr 22 Gateway Protocol • Mix and incubate for 60’ @ 25 oC Clonase reaction buffer 4 mL • Add proteinase K and incubate for 10’ Destination Vector 300 ng at 37 oC Entry Clone 100 ng • Transfer to E. coli Clonase Enzyme mix 4 mL (competent) DH5 Total volume 20 mL cells • Express for 60’ and plate on LB-Amp Ingredients • • • • • Lecture 3.0 23 Expression/Cloning -Which Protocols? • Molecular Cloning 3rd Edition (Sambrook and Maniatis / Russell) – http://www.molecularcloning.com • Molecular Biology Protocols – http://micro.nwfsc.noaa.gov/protocols/ • Molecular Biology Shortcuts – http://highveld.com/f/fprotocols.html • Protocols Online – http://www.protocol-online.org/ Lecture 3.0 24 Expression Questions • • • • • • • • Which host cell system? Which expression vector? Which cloning/expression protocols? Is it membrane or water soluble? Is it single domain or multi-domain? How soluble and how stable? Where will this protein be found? How to purify & how to identify? Lecture 3.0 25 Membrane or Water Soluble? Lecture 3.0 26 Membrane or Water Soluble? • Most protein scientists prefer to work with water soluble proteins or domains • Membrane proteins are very difficult to clone, express and purify and special techniques must be used • Potential problems can be avoided by knowing whether the protein contains one or more membrane spanning helices and where these helices are located (cleaved?) Lecture 3.0 27 Predicting via Hydrophobicity Bacteriorhodopsin 4 2 OmpA 3 1.5 2 1 0.5 1 0 0 -0.5 1 -1 -1 -2 -1.5 -3 -2 Bacteriorhodoposin Lecture 3.0 OmpA 28 Membrane Helix Prediction • Neural Network and HMM methods now claim >80% accuracy • PredictProtein (PHDhtm) – http://cubic.bioc.columbia.edu/predictprotein/ • TMpred – http://www.ch.embnet.org/software/TMPRED_form .html • TMHMM – http://www.cbs.dtu.dk/services/TMHMM-2.0/ Lecture 3.0 29 TMPred (Principles) Table 6 Protein Family Cytokine/growth factor receptors (EGF, interleukin, Insulin receptors) G-coupled receptors (rhodopsin, bacteriorhodopsin etc.) Extracellular activated gated channels (Glutamate, GABA, ACh sensitive) Intracellular activated gated channels 5) photosynthetic proteins (H chain) 6) photosynthetic proteins (L chain) 7) porins 8) microsomal cytochrome p450 9) cytochrome b 10) Fo ATPases Lecture 3.0 No. of Membrane Segments 1 (helix) 7 (helices) 4 (helices) 6 (helices) 5 (helices) 5 (helices) 17 (b-strands) 1 (helix) 1 (helix) 4 (helices) 30 TMHMM Lecture 3.0 31 PredictProtein Lecture 3.0 32 Expression Questions • • • • • • • • Which host cell system? Which expression vector? Which cloning/expression protocols? Is it membrane or water soluble? Is it single domain or multi-domain? How soluble and how stable? Where will this protein be found? How to purify & how to identify? Lecture 3.0 33 Single Domain or MultiDomain? Lecture 3.0 34 Modular Protein Domains BH PDZ FYVE PH DED DEATH SH3 1433 WW Lecture 3.0 FHA PTB SH2 35 Single Domain or MultiDomain? • Many eukaryotic proteins are multi-domain • Size is a good indicator (roughly 1 domain for every 15 kD) • Small domains behave better (Xray & NMR) • Limited proteolysis allows experimental identification of domains prior to structure determination by NMR or X-ray – digestion followed by HPLC or MS analysis to detect fragments > 10 kD Lecture 3.0 36 Domain Prediction • Domain Prediction (PredictProtein-GLOBE) – http://cubic.bioc.columbia.edu/predictprotein • BLAST alignments can be used to detect or predict the presence of domains by sequence homology • Protein domains can also be predicted using CDD (Conserved Domain Database) at http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml Lecture 3.0 37 Lecture 3.0 38 Lecture 3.0 39 Expression Questions • • • • • • • • Which host cell system? Which expression vector? Which cloning/expression protocols? Is it membrane or water soluble? Is it single domain or multi-domain? How soluble and how stable? Where will this protein be found? How to purify & how to identify? Lecture 3.0 40 Predicting Solubility • Even if a protein is identified to be a nonmembrane protein this does not necessarily indicate it will be soluble • Solubility depends on many factors – – – – – size (smaller ones are more soluble) hydrophobicity (average and local hphob) 3D structure and ligand interactions overall charge, predicted accessibility distribution and frequency of amino acids Lecture 3.0 41 Predicting Solubility • Solvent accessibility prediction – PredictProtein (PHDacc) – http://cubic.bioc.columbia.edu/predictprotein/ • Protein property/scale prediction – EXPASY ProtScale – http://www.expasy.ch/cgi-bin/protscale.pl • PepTool – www.biotools.com Lecture 3.0 42 Accessible Surface Area Reentrant Surface Solvent Probe Accessible Surface Van der Waals Surface Lecture 3.0 43 Score Predicted Accessibility 3 32 1 20 -1 1 -2 -3 0 -4 1 Lecture 3.0 51 101 151 201 251 301 44 Buried Surface Area (BASA) & Fractional Burial (FB) • For an average protein • ASA (NP) = 0.35 x BASA • ASA (P) = 0.61 x BASA • ASA (+/-) = 0.04 x BASA • BASA can be estimated from a protein’s amino acid composition BASA = S AAi x FBi Lecture 3.0 Table 9 Amino Acid Residue Fraction Buried Residue frac. bur. Residue frac. bur. A C D E F G H I K L 0.38 0.45 0.15 0.18 0.5 0.36 0.17 0.6 0.03 0.45 M N P Q R S T V W Y 0.4 0.12 0.18 0.07 0.01 0.22 0.23 0.54 0.27 0.15 45 ProtScale Lecture 3.0 46 ProtScale Lecture 3.0 47 Solubility (PepTool) • Average Hydrophobicity AH = S AAi x Hi • Hydrophobic Ratio RH = S H(-)/S H(+) • Hydrophobic % Ratio RHP = %philic/%phobic • Linear Charge Density LIND=(K+R+D+E+H+2)/# • Solubility SOL=RH + LIND 0.05AH Lecture 3.0 • Average AH = 2.5 +/- 2.5 Insol > 0.1 Unstrc < -6 • Average RH = 1.2 +/- 0.4 Insol < 0.8 Unstrc > 1.9 • Average RHP = 0.9 +/- 0.2 Insol < 0.7 Unstrc > 1.4 • Average LIND = 0.25 Insol < 0.2 Unstrc > 0.4 • Average SOL = 1.6 +/- 0.5 Insol < 1.1 Unstrc > 2.5 48 Structural Proteomics and Solubility Prediction • Global efforts have led to the cloning and attempted expression of more than 5000 water soluble proteins • Data contained on databases such as TargetDB allow correlations to be developed between sequence and expression levels and solubility • Excellent opportunity to used data mining to find “rules” to predict protein solubility Lecture 3.0 49 Lecture 3.0 50 Binary Decision Trees • Used to partition or classify data that is not linearly separable • Unknown objects are classified by “traversing” the tree • Traversing is accomplished by performing tests at each node, direction of traversal determined by results of the test • Decision trees can be trained (test threshold cutoff, test order, architecture) Lecture 3.0 51 Binary Decision Trees # not forming crystals Lecture 3.0 # forming crystals 52 Predicting Protein Solubility 1) Residue frequency [ACDEFGHIKLMNPQRSTVWY] 2) Grouped residue frequency {[KR],[NR],[DE],[ST] [LIM],[FWY],[HKR],[AVILM],[DENQ],[GAVL],[SCTM]} 3) Predicted % secondary structure [a,b,c] 4) Presence of signal sequence 5) Length of polypeptide 6) Number of residues in low complexity region (L,S) 7) Normalized low complexity value (SEG/Len) 8) Maximum hydrophobicity value 9) Length of maximum hydrophobic region Lecture 3.0 53 Solubility Decision Tree Size of black oval = % that are soluble Lecture 3.0 54 Binary Decision Trees • Have been used to predict protein solubility and protein crystallization • Somewhat similar to self-organizing feature maps (SOFM) • Bertone P, Kluger Y, Lan N, Zheng D, Christendat D, Yee A, Edwards AM, Arrowsmith CH, Montelione GT, Gerstein M. Nucleic Acids Res 2001 1;29(13):288498 Lecture 3.0 55 Predicting Stability • Even if a protein expresses and remains soluble it may turn out to be quite unstable (easily proteolyzed) • Proteins that are rich in Proline (P), Glutamic acid (E), Serine (S) and Threonine (T) or which have regions that are rich in these amino acids (PEST sequences) tend to have half lives of less than 2 hours Lecture 3.0 56 PEST Finder http://www.at.embnet.org/embnet/tools/bio/PESTfind/ Lecture 3.0 57 Expression Questions • • • • • • • • Which host cell system? Which expression vector? Which cloning/expression protocols? Is it membrane or water soluble? Is it single domain or multi-domain? How soluble and how stable? Where will this protein be found? How to purify & how to identify? Lecture 3.0 58 Protein Localization • Is it exported? Does it go to the nucleus? Does it go through the ER? Does it localize to mitochondria? Chloroplasts? Does it go to the membrane? How do you tell? • Eukaryotic signal sequences are usually incompatible with prokaryotic signal sequences so expressing eukaryotic proteins in bacteria can lead to problems Lecture 3.0 59 Location Prediction http://psort.nibb.ac.jp Lecture 3.0 60 Proteome Analyst http://www.cs.ualberta.ca/~bioinfo/PA/Sub/ Lecture 3.0 61 PSORT-B (bacteria) http://www.psort.org/psortb/index.html Lecture 3.0 62 Location Prediction http://www.cbs.dtu.dk/services/TargetP/#submission Lecture 3.0 63 Other Sites or Modifications? • Phosphorylation – NetPhos http://cbs.dtu.dk/services/NetPhos/ • O-Glycosylation – NetOGlyc http:/cbs.dtu.dk/services/NetOGlyc/ • Coil-Coil Dimerization domains – www.ch.embnet.org/software/COILS_form.html • Tyrosine Sulfation – http://ca.expasy.org/tools/sulfinator/ Lecture 3.0 64 NetPhos 2.0 Lecture 3.0 65 Expression Questions • • • • • • • • Which host cell system? Which expression vector? Which cloning/expression protocols? Is it membrane or water soluble? Is it single domain or multi-domain? How soluble and how stable? Where will this protein be found? How to purify & how to identify? Lecture 3.0 66 Finding and Identifying Your Protein Lecture 3.0 67 Isoelectric Point • The pH at which protein has charge=0 • Q = S Ni/(1 + 10pH-pKi) pKa Values for Ionizable Amno Acids Residue C D E Lecture 3.0 pKa 10.28 3.65 4.25 Residue H K R pKa 6 10.53 12.43 68 Isoelectric Point & MW Calculation Lecture 3.0 69 More Help? • • • • • • • http://www.abrf.org http://www.abrf.org.JBT/JBTindex.html http://www.BioTechniques.com http://expasy.ch/alinks.html http://www.neehow.org/wonderful/protocols http://research.newfsc.noaa.gov/protocols.html http://www.horizonpress.com/gateway/protocol s.html Lecture 3.0 70 Bioinformatics & Structural Proteomics • Key to identifying targets • Key to reducing time and material wastage in protein expression/purification steps • Key to tracking and communicating target progression (multi-lab LIMS) • Key to reducing redundancy and duplication by other X-ray or NMR structure labs (TargetDB, SPINE) Lecture 3.0 71 TargetDB Lecture 3.0 http://targetdb.pdb.org/ 72 Structural Proteomics - Status • • • • • • • • 20 registered centres (~30 organisms) 82700 targets have been selected 52705 targets have been cloned 29855 targets have been expressed 12311 targets are soluble 1493 X-ray structures determined 502 NMR structures determined 1743 Structures deposited in PDB Lecture 3.0 73 Structural Proteomics - Status • • • • • • • 543 structures deposited by Riken 265 structures deposited by Mid-West 187 structures deposited by North-East 179 structures deposited by New York 178 structures deposited by JCSG (UCSD) 52 structures deposited by Berkeley 31 structures deposited by Montreal/Kingston Lecture 3.0 74 Protein Expression in E. coli good promising unfolded poor precipitated Lecture 3.0 75 Proc. Natl. Acad. Sci. USA, Vol. 99,1825-1830, 2002 Protein Expression in E. coli Cloned (517 total) expressed (85%) soluble (68%) M. th.= Methanobacter thermoautotrophicum E. coli = Escherichia coli S. ce. = Saccharomyces cerevisae Myx. = Myxoma virus T. ma. = Thermotoga maritima Lecture 3.0 76 X-ray vs. NMR Results for Methanobacter Lecture 3.0 77 Conclusions • The success of proteomics (structural, functional, expressional) hinges almost entirely on successful protein production and expression • Bioinformatics (web databases, servers, data mining tools, NN’s, HMM’s) can and does play an increasingly important role in optimizing or improving protein expression and coordinating large scale proteomics efforts Lecture 3.0 78