* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Designing Minor Groove Binding Drugs
DNA barcoding wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Biochemistry wikipedia , lookup
Synthetic biology wikipedia , lookup
Chemical biology wikipedia , lookup
Restriction enzyme wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Transformation (genetics) wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Biosynthesis wikipedia , lookup
Point mutation wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Genome editing wikipedia , lookup
DNA vaccination wikipedia , lookup
Molecular cloning wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
History of genetic engineering wikipedia , lookup
Non-coding DNA wikipedia , lookup
DNA supercoil wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Designing Minor Groove Binding Drugs
Vonetta Edwards, Kathy Goodson, Habi Mojidi and Sarah Mae Sucayan
Biochemistry 674, University of Maryland, College Park, MD
Introduction
Current Strategies
Factors in Recognizing the Minor Groove
Transcription or replication of DNA only occurs after a signal has been received, usually in the form
of a protein that binds to a particular region of the DNA. If a small artificial protein can be developed
that would mimic the binding strength and specificity of the natural regulatory protein, then DNA
function can be artificially modulated, inhibited or activated. Drugs may bind irreversibly, or reversibly
in which non-covalent bonds are formed. The latter is preferred when developing drugs.1-3
Electrostatics
• A/T sequences (Figure 2A) have a higher negative
potential while G/C sequences (Figure 2B) have
higher positive potential
• H-bonding to base edges in the minor groove can
be used to distinguish between A:T and G:C base
pairs
• A:T is distinguished from a T:A base pair through
indirect read out
• Minor groove binders – are crescent shaped, bind via Van der Waals interactions and hydrogen
bonds and have a preference for AT rich sequences. e.g. distamycin, Hoescht 33258.
• Intercalators – consist of planar heterocyclic/chromopore groups that stack between adjacent
DNA bases and also have hydrogen interactions. Most prefer GC rich regions (bleomycin).
A
Categories of Sequence Specific DNA Binding Drugs
B
One of the first chemical approaches to targeting
double-stranded DNA used oligonucleotides linked
to intercalators to bind in the major groove of DNA
and form a sequence specific triple helix.4
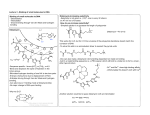
Figure 74: Triple Helix Model
Figure 25: Electrostatic potential surface of
dsDNA crystal structures.
Another strategy in improving sequence
specificity of minor groove binding drugs is the
use of polyamides. These group of molecules
contain combinations of three different aromatic
amino acids (hydroxypyrrole, imidazole, pyrrole),
which uniquely bind with each of the WatsonCrick base pairs.(see Figure 9).4
Structure
• Triple Helix forming molecules
Major
groove
• Synthetic Polyamides
• Zn++ Finger proteins
Width
Depth
Major
Groove
11.6Å
8.5Å
Minor
Groove
6.0Å
8.2Å
Major
groove
Minor
groove
Figure 14:Interaction of the different combinations of
amides with specific WC base pairs.
Minor
groove
Figure 84: Hairpin Polyamide Model
Figure 3: Left: Groove dimensions5. Right: WC base pairs.7
Biological Effects of DNA Binding Drugs
Transcription factors – The most effective inhibitors bind to both the major and minor grooves and
allow ‘threading’ which keeps the DNA inaccessible to transcriptions factors.1
Topoisomerases – “cleavable complex” poisons ,such as quinolones, stabilize covalent enzyme-DNA
complexes thus prevent resealing of DNA ends. This converts type II enzymes into a cellular
poison..“catalytic inhibitors,” such as coumermycin, inhibit essential ATP hydrolysis needed for
topoisomerase function.3
• DNA binding drugs are generally flat and small, which makes them fit well into the
minor groove.
• Groove width varies with sequence; A/T rich tracks tend to make the groove width
narrower.
• The convex shape of the minor groove floor complements the typical shapes of minor
groove binding drugs
• A/T sequences result in a smooth convex curve whereas G/C sequences have “little”
bumps due to the 2-amino groups of guanine.
Figure 9 :Molecules such as Pyrole-imidazole polyamides are linear Beta-alanine
linked polyamides that recognize a large range of DNA.12
Initial Probe Studies
Conclusion and Future Work
Beginning with the work of Galas and Schmitz,
methods of DNA footprinting1 with DNase I allowed
for determination of the location of molecular binding
sites on DNA.8 A synthetic cleavage agent
methidiumpropyl-EDTA (MPE) is comprised of
methidium, a DNA intercalator, which is covalently
bound to the metal chelator EDTA. This agent was later
adopted for use in footprinting due to its smaller size
and its preferable mode of activation.9
5’
3’
Fe
Fe
Figure 411: MPE
The mode of cleavage of
EDTA•Fe(II) is through the use of
a non-specific hydroxyl radical,
thus cleavage is attributed to the
binding ligand. In experiments by
Younguist and Dervan, EDTA was
attached to either the amino or
carboxyl end of tri-, tetra-, penta-,
and
hexa(tris-Nmethylpyrrolecarboxamide)s. The
result of experimentation was the
outline of an n+1 binding motif,
where n is the number of
amides.10
5’
3’
5’
3’
High Resolution Gel
TA
AT
TA
CG
TA
TA
TA
AT
AT
CG
GC
TA
AT
GC
5’ 3’
Figure 6:EDTA•Fe(II) affinity cleavage model reflecting the asymmetric DNA cleavage
pattern seen in the minor groove of B-DNA. The frequency of cleavage is represented
by the lengths of the arrows. Figure adapted from Youngquist and Dervan.10
Figure 511: Amino and carboxyl labeled tris-Nmethylpyrrolecarboxamide
DNA binding molecules have various affinities for specific regions of DNA. Synthetic analogs of
the AT-selective minor groove-binding ligands13 created the foundation for synthetic DNA binding
drugs. Sequence specificity of DNA binding drugs will provide insight into drug design that will target
genes and be used as a class of potential therapeutics against unknown biological weapons and
personalized medicines.14 Currently, various clinical trials of genetic therapies are in progress to find
effective, safe designer drugs to target various harmful genetic conditions. Once some of the
conditions have been met this will lead to the possible eradication of some genetic disorders.
References
1. Bassi, L.; Palitti, F., Anti-topoisomerase drugs as potent inducers of chromosomal aberrations. Genetics and Molecular Biology 2000, 23, (4), 1065-1069.
2. Gambari, R.; Feriotto, G.; Rutigliano, C.; Bianchi, N.; Mischiati, C., Biospecific interaction analysis (BIA) of low-molecular weight DNA-binding
drugs. Journal of Pharmacology and Experimental Therapeutics 2000, 294, (1), 370-377.
3. Welch, J. J.; Rauscher, F. J.; Beerman, T. A., Targeting DNA-Binding Drugs to Sequence-Specific Transcription Factor DNA Complexes
Differential-Effects of Intercalating and Minor-Groove Binding-Drugs. Journal of Biological Chemistry 1994, 269, (49), 31051-31058.
4. Uil, T. G.; Haisma, H. J.; Rots, M. G., Therapeutic modulation of endogenous gene function by agents with designed DNA-sequence specificities.
Nucl. Acids Res. 2003, 31, (21), 6064-6078.
5. Neidle, S., DNA minor-groove recognition by small molecules. Natural Product Reports 2001, 18, (3), 291-309.
6. Moser, H. E.; Dervan, P. B., Sequence-specific cleavage of double helical DNA by triple helix formation. Science 1987, 238, (4827), 645-650.
7. Dickerson, R. E., In Oxford Handbook of Nucleic Acid Structure, Neidle, S., Ed. Oxford University Press: 1999; pp 145-197.
8. Galas, D. J.; Schmitz, A., DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res 1978, 5, (9),
3157-70.
9. Van Dyke, M. W.; Dervan, P. B., Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on
DNA. Nucleic Acids Res 1983, 11, (16), 5555-67.
10. Youngquist, R. S.; Dervan, P. B., Sequence-Specific Recognition of B-DNA by Oligo(N-methylpyrrolecarboxamide)s. PNAS 1985, 82, (9), 25652569.
11. Dervan, P. B., Design of sequence-specific DNA-binding molecules. Science 1986, 232, (4749), 464-71.
12. Burnett, R.; Melander, C.; Puckett, J. W.; Son, L. S.; Wells, R. D.; Dervan, P. B.; Gottesfeld, J. M., DNA sequence-specific polyamides alleviate
transcription inhibition associated with long GAA{middle dot}TTC repeats in Friedreich's ataxia. PNAS 2006, 103, (31), 11497-11502.
13. Schaal, T. D.; Mallet, W. G.; McMinn, D. L.; Nguyen, N. V.; Sopko, M. M.; John, S.; Parekh, B. S., Inhibition of human papilloma virus E2 DNA
binding protein by covalently linked polyamides. Nucleic Acids Research 2003, 31, (4), 1282-1291.