* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Gregor Mendel - BEHS Science
Pharmacogenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Behavioural genetics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene desert wikipedia , lookup
Gene therapy wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genome (book) wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Gene nomenclature wikipedia , lookup
Genetically modified crops wikipedia , lookup
Gene expression programming wikipedia , lookup
X-inactivation wikipedia , lookup
Copy-number variation wikipedia , lookup
Public health genomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Selective breeding wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Microevolution wikipedia , lookup
Designer baby wikipedia , lookup
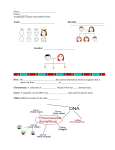
Mendelian Genetics Chapter 14 Gregor Mendel 1850s, Mendel studied inheritance in pea plants by breeding them Peas are good because they have distinctive traits, male and female parts, and are easy to manipulate He crossed true breeding with other true breeding as well as with hybrids Gregor Mendel P = parental generation F1 = First filial generation (P X P) F2 = Second filial (F1 X F1) True breeding (homozygous) = plants that only breed to produce one phenotype Hybrid (heterozygous) = results of crosses between two plants that breed true for different phenotypes for the same trait Gregor Mendel Genotype = genetic makeup of an organism – Sequence of the gene Phenotype = expressed genotype – Visible or measurable traits – based upon genotype Gregor Mendel Gregor Mendel He crossed a true breeding purpleflowered plant with a true breeding white-flowered plant All of the F1 generation was purple Gregor Mendel He then crossed two F1’s with each other The results (F2’s) were roughly 3 purples to 1 whites White trait was absent in F1’s but reappeared in the F2’s Gregor Mendel The reappearance of the white trait meant that it was not altered in the F1, just “hidden” Mendel did the same crosses, and got similar results, with other traits of the pea plant Gregor Mendel Four ideas spawn from Mendel’s work: 1. Alleles account for different traits 2. Organisms inherit two alleles, one from each parent (may be same or different) 3. If alleles are different, then one is dominant over the other 4. The two alleles segregate (law of segregation) during gamete formation Punnett Square Predicts the statistical results of a cross Punnett Square A test cross, breeding a homozygous recessive with a dominant phenotype, but unknown genotype, can determine the identity of the unknown allele Punnett Square One Factor Crosses = crosses with only one trait – Monohybrid crosses = crossing one heterozygote with another Two Factor Crosses = crosses with two traits – Dihybrid crosses = crossing one organism that is heterozygous for both traits with another that is heterozygous for both traits Punnett Square In monohybrid crosses, he noticed that genotypic and phenotypic ratios could be predicted – Genotypic ratio = 1:2:1 – Phenotypic ratio = 3:1 Punnett Square Example of a dihybrid cross: – Crossing one plant that is heterozygous for seed shape and yellow seed coat (RrYy) with another plant that is also heterozygous for both traits (RrYy) – RrYy X RrYy Dihybrid results point to the law of independent assortment Independent Assortment In our example (RrYy X RrYy), both parents can produce four possible gametes – RY – Ry – rY – Ry Since both parents have four possible gametes, there are 16 possible combinations for fertilization Independent Assortment Independent Assortment These possible combinations produce a genotypic ratio of 1:2:1:2:4:2:1:2:1 It also produces a phenotypic ratio of 9:3:3:1 The law of independent assortment simply means that during gamete formation, allelic pairs that code for different traits assort independent of each other Dominance Mendel was very lucky to have studied peas – They follow simple dominance laws – Not all cells follow the simple laws of dominance vs. recessiveness Variations Incomplete dominance = neither allele is shown in its dominant form; an intermediate phenotype is shown – Genotypic and phenotypic ratios are the same (1:2:1) Variations Codominance = heterozygote expresses a phenotype that is distinct from and not intermediate between those of the two homozygotes Ex. Human AB blood type Variations Codominance Variations Multiple alleles – gene exists as more than two alleles in the population – Rabbit coat color gene has 4 alleles: C, c, cch & ch – 5 phenotypes – 10 genotypes Variations Pleiotropy – one gene affects more than one phenotypic characteristic – Ex: sickle-cell disease affects much more than just the overall conformation of the hemoglobin protein Variations Epistasis – one gene at one locus affects the phenotype of another gene at another locus – Ex: mice coat color depends on two genes – One, the epistatic gene, determines whether or not pigment will be deposited in hair • Presence (C) is dominant to absence (c) – The second determines whether the pigment to be deposited is black (B) or brown (b) – An individual that is cc has a white (albino) coat regardless of the genotype of the second gene Variations – epistasis A cross between two black mice that are heterozygous (BbCc) will follow the law of independent assortment However, unlike the 9:3:3:1 offspring ratio of an normal Mendelian experiment, the ratio is 9 black, 3 brown, and 4 white Variations Polygenic – one phenotype determined by the combined effect of more than one gene – Ex: human skin color, eye color, height Variations Variations Multifactorial Traits – determined by the combined effect of one or more genes plus the environment – Ex: heart disease, body weight, intelligence Pedigrees Autosomal Recessive Traits: – located on non-sex chromosomes – affects males and females – parents must be carriers or affected – affected individuals are homozygous recessive Ex: Albinism, Cystic fibrosis, Phenylketonuria, Sickle cell disease Autosomal Recessive Pedigrees Autosomal Dominant Traits: – located on non-sex chromosomes – affects males and females – at least one parent is affected – does not skip generations – affected individuals are homozygous dominant or heterozygous Ex. Achondroplasia, Huntington disease, Lactose intolerance, Polydactyly Autosomal Dominant