* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download RNA Splicing
Metagenomics wikipedia , lookup
Transposable element wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Hammerhead ribozyme wikipedia , lookup
Protein moonlighting wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Non-coding DNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Human genome wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
RNA interference wikipedia , lookup
Transfer RNA wikipedia , lookup
Messenger RNA wikipedia , lookup
Polyadenylation wikipedia , lookup
Deoxyribozyme wikipedia , lookup
RNA silencing wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Non-coding RNA wikipedia , lookup
History of RNA biology wikipedia , lookup
RNA-binding protein wikipedia , lookup
Epitranscriptome wikipedia , lookup
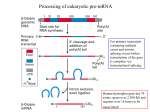
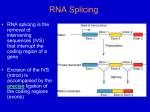
Interrupted Genes • Prokaryotes – continuous gene(uninterrupted) • Eukaryotes – gene is interrupted with non-coding sequences (introns) RNA splicing – the removal of these introns while joining the rest • Terminology Exons – sequences represented in mature RNA (A gene starts and ends with exons that correspond to the 5’ and 3’ ends of RNA) Introns – Intervening sequences that are removed when the primary transcript is processed to the mature RNA DNA TRANSCRIPT(RNA COPY) MATURE RNA • The mechanism excludes any splicing together of sequences representing different alleles • A typical mammalian gene has 7 to 8 exons spread out over ~16 kb . The exons are relatively short(~100 – 200 bp) and introns relatively long(>1 kb) • So, gene is interrupted while mRNA(~2.2 kb) is uninterrupted , which requires the primary transcript(pre-mRNA) to be processed. • Nuclear RNA(including pre-mRNA) - much larger than mRNA - very unstable - much greater sequence complexity - known as hnRNA(heterogenous nuclear RNA) • hnRNP – ribonuclear protein ; the physical form of hnRNA,which is bound to proteins ; Has the form of beads connected by a fiber. • Splicing and other post transcriptional modifications take place in the nucleus SPLICING IS OF SEVERAL TYPES • In higher eukaryotes – introns removed by a system that recognizes only short consensus sequences conserved at exon-intron boundaries and within the intron. - Requires spliceosomes (large splicing apparatus) - Mechanism involves transesterifications - Catalytic center includes RNA and proteins. • Autonomous splicing – of introns by certain RNAs - 2 types of introns – distinguished by 20 and 30 structures - Mechanism – transesterification - Catalytic agent – RNA (catalytic RNA) • Splicing of yeast tRNA – accomplished by enzymes that use cleavage and ligation. SPLICE JUNCTIONS (splice sites) • The two exon-intron boundaries that include the sites of breakage and reunion. ie, the junction between exons and introns. • There is no extensive homology or complementarity between 2 ends of an intron. But there are well conserved,short,consensus sequences. • High conservation is found only immediately within the intron at the presumed junctions. • GT-AG rule – ie., GU-AG rule in pre-mRNA - An intron starts with dinucleotide GU and ends in AG - called 5’ and 3’ splice sites resp. SPLICE JUNCTIONS ARE READ IN PAIRS • In an mRNA, introns are multiple and long • Appropriate 5’ and 3’ sites should be paired - It could be an intrinsic property of RNA to connect the sites at the ends of a particular intron - Splicing could follow rules that ensure a 5’ site is always paired to a following 3’ site • In principle any 5’ splice site can be connected to any 3’ splice site. So,there are preferred pathways that ensure right splicing. • The conformation of the RNA influences the accessibility of the splice sites. As particular introns are removed,the conformation changes and new pairs of splice sites become available. • So , the splicing reaction does not proceed sequentially along the precursor RNA. SPLICING PROCEEDS THROUGH A LARIAT • Splicing is independent of transcription or other post – transcriptional modifications,yet occur co-ordinated. • In vivo, the exons are not released as free molecules during splicing,but remain held together by the splicing apparatus. • Splicing requires the 5’ and 3’ splice sites and a branch site just upstream of the 3’ splice site. • Steps in splicing - A cut is made at the 5’ splice site, separating the left exon and the right intron-exon molecule SPLICING PROCEEDS THROUGH A LARIAT - The left exon becomes linear - The right intron-exon molecule form a lariat by forming a 5’-2’ bond between 5’ terminus and the target base ‘A’ called the branch site - The 3’ splice site is then cut releasing free intron in the lariat form - The right exon is ligated (spliced) to the left exon - The lariat is then debranched to give a linear excised intron which is rapidly degraded • The branch site plays an important role in identifying the 3’ splice site. The consensus is highly conserved in yeast as UACUAAC. • The branch site is not well conserved in higher eukaryotes, but has a preference for bases at each position and retains the target A nucleotide. • The branch site lies 18 to 40 nucleotides upstream of the 3’ splice site. • The lariat formation is effected by transesterification - First, a nucleophilic attack by the 2’-OH of the invariant A on the 5’ splice site - Second, the free 3’-OH of the exon that was released , now attacks the bond at the 3’ splice site THE SPLICING APPARATUS • Contains both proteins and RNAs ; Splicing occurs only after all components are sequentially assembled on the pre-mRNA • The small RNAs are found both in nucleus and cytoplasm of eukaryotic cells In nucleus – small nuclear RNAs (snRNAs) In cytoplasm – small cytoplasmic RNAs (scRNAs) In nucleolus – snoRNAs • They exist as ribonucleoprotein particles snRNPs and scRNPs (known colloq. as snurps and scyrps) • Spliceosome – large particulate complex formed of snRNPs involved in splicing and many additional proteins - It comprises a 50S to 60S RNP particle • Like the ribosome, the spliceosome depends on RNARNA, protein-RNA and protein-protein interactions. • The 5 snRNPs involved in splicing are U1, U2, U5, U4 and U6 . Each snRNP contains a single snRNA and several(>20) proteins. U4 and U6 are usually found as a single U4/U6 particle. SPLICEOSOME MACHINERY • Before any irreversible change is made to the RNA, all of the splicing components are assembled and have ensured that the splice sites are available. • Splicing is divided into 2 stages a) The 5’ splice site, branch sequence and adjacent pyrimidine tract are recognised.The spliceosome complex is assembled b) Structure of transcript is changed by cleavage and ligation. Components of the complex are released or reorganised as it proceeds through the reactions. • Binding of U1 snRNP to the 5’ splice site is the first step in splicing. ie.,one of its proteins,U1-70k interacts with protein ASF/SF2(an SR class general splicing factor) causing U1 snRNA to base pair with the 5’ site by a single stranded region at 5’ terminus (4 to 6 bases complementary with splice site). • Complementarity between U1 snRNA and 5’ splice site is necessary for splicing, with pairing stabilized by proteins of U1 snRNP. [SR proteins – imp. group of splicing factors & regulators - Take their name from Ser-Arg rich region with variable length. They interact each other via these regions. They bind RNA/connects U2AF to U1. They are essential part of spliceosome,forming a framework on RNA substrate] • The first complex formed during splicing is the E (early presplicing) complex – it contains U1 snRNP,U2AF(a splicing factor) and some SR proteins. • The formation of E complex identifies a pre-mRNA as a substrate for formation of splicing complex and is hence also called the commitment complex. • In the E complex, U2AF is bound to the region between the branch site and the 3' splice site. In most organisms, it has a large subunit (U2AF65) that contacts a pyrimidine tract downstream of the branch site; a small subunit (V2AF35) directly contacts the dinucleotide AG at the 3' splice site. • Another splicing factor, called SF1in mammals and BBP in yeast. connects V2AF/Mud2 to the U1 snRNP bound at the 5' splice site. Complex formation is enhanced by the cooperative reactions of the two proteins; SF 1 and U2AF (or BBP and Mud2) bind together to the RNA substrate -1 Ox more effectively than either alone. This interaction is probably responsible for making the first connection between the two splice sites across the intron. • The E complex is converted to the A complex when U2 snRNP binds to the branch site. Both UI snRNP and U2AF/Mud2 are needed for U2 binding. The U2 snRNA includes sequences complementary to the branch site. • A sequence near the 5’ end of the snRNA base pairs with the branch sequence in the intron. Several proteins of the U2 snRNP are bound to the substrate RNA just upstream of the branch site. • The binding of U2 snRNP requires ATP hydrolysis and commits a pre-mRNA to the splicing pathway by generating A presplicing complex. Formation of E complex • Intron definition – The two splice sites are recognised without requiring any sequences outside of the intron. • The SR proteins may enable U2AFlU2 snRNP to bind in vitro in the absence of UI, raising the possibility that there could be a U1-independent pathway for splicing • Exon definition – When introns are long and splice sites are weak ; sequences downstream of the intron itself are required ; The 3‘ splice site is recognized as part of a complex that forms across the next exon. though, in which the next 5' splice site is also bound by UI snRNA. This UI snRNA is connected by SR proteins to the U2AF at the pyrimidine tract. 5 snRNPs Form the Spliceosome • The snRNPs and factors associate with E complex in a defined order. • B1 complex – formed when a trimer U5 and U4/U6 binds to A complex(U1 and U2 snRNPs) This is the spliceosome complex – has all components needed for splicing. • B2 complex – formed when U1 snRNA is released,other components,esp U6 comes into juxtaposition with 5’ splice site, and U5 shifts to the vicinity of intron sequences. • The role of U4 snRNA may be to sequester U6 snRNA until it is needed. So U4 is released with hydrolysis of ATP ,triggering catalytic reaction. • When U4 is released,the region of U6 initially base paired with U4 now is free. The first part of it pairs with U2; the second part forms an intramolecular hairpin. • Thus several pairing reactions between snRNAs and the substrate RNA occur in the course of splicing. • U6 snRNA is not used up in a splicing reaction and at completion must be released from U2 so that it can reform the duplex structure with U4 to undertake another cycle of splicing. ALTERNATE SPLICING APPARATUS • In human genome,more than 98% introns are GU-AG . Less than 1% are GC-AG About 0.1 % are AU-AC type. • These introns required an alternate splicing apparatus that comprise the U12 spliceosome,containing U11 , U12, a U5 variant and the U4atac and U6atac snRNAs. • The splicing reaction is essentially similar to that of GU – AG introns. • Some GU-AG introns may also be spliced by the U12 spliceosome and vice-versa. • The two types of introns co-exist in a variety of genomes and may even be found in the same gene. AUTOSPLICING • Introns in protein coding genes are generally of 3 classes * nuclear pre-mRNA introns * Group I introns * Group II introns • Nuclear pre-mRNA introns are identified by the presence of GU-AG base sequence • Group I and II introns are found in organelles and bacteria. Group I introns are more common.Each can be folded into a typical type of secondary structure. • They have the ability to excise themselves from an RNA – ie autosplicing. In vivo,proteins are required to assist folding. • All 3 classes of introns are excised by two successive transesterification reactions. • There are parallels between group II introns and pre-mRNA splicing. Group II mitochondrial introns are excised by the same mechanism as nuclear pre-mRNAs via a lariat that is held together by a 5'-2' bond • The ability of group II introns to remove themselves by an autocatalytic splicing event stands in great contrast to the requirement of nuclear introns for a complex splicing apparatus. • The snRNAs of the spliceosome as compensating for the lack of sequence information in the intron, and providing the information required to form particular structures in RNA and may have evolved from the autocatalytic system. • Thus the snRNAs may undergo reactions with the premRNA substrate, and with one another, that have substituted for the series of conformational changes that occur in RNAs that splice by group II mechanisms. • These changes have relieved the substrate pre-mRNA of the obligation to carry the sequences needed to sponsor the reaction. As the splicing apparatus has become more complex (and as the number of potential substrates has increased), proteins have played a more important role. ALTERNATIVE SPLICING • When an interrupted gene is transcribed into an RNA that gives rise to a single type of spliced mRNA, there is no ambiguity in assignment of exons and introns. • But when a single gene gives rise to more than one mRNA sequence,it follows an alternative splicing pattern. • In some cases, the ultimate pattern of expression is dictated by the primary transcript, because the use of different startpoints or the generation of alternative 3' ends alters the pattern of splicing. • In other cases, a single primary transcript is spliced in more than one way, and internal exons are substituted, added, or deleted. • In some cases, the multiple products all are made in the same cell, but in others the process is regulated so that particular splicing patterns occur only under particular conditions. • There is an ASF(Alternative Splicing Factor) which is same as that of the SF2 splicing factor.Both are RNA binding proteins in the SR family. • sxl > tra • When a pre-mRNA has more than one 5' splice site preceding a single 3' splice site, increased concentrations of ASF/SF2 promote use of the 5' site nearest to the 3' site at the expense of the other site. This effect of ASF/SF2 can be counteracted by another splicing factor, SF5. • Alternative splicing may also be influenced by repression of one site. TRANS-SPLICING REACTIONS • In genetic terms, splicing occurs only in cis. This means that only sequences on the same molecule ofRNA can be spliced together. • Very rare and observed in vitro usually. • Seen in vivo,in some special situations. • When splicing occurs, a 5'-2' link forms by the usual reaction between the GU of the 5‘ intron and the branch sequence near the AG of the 3' intron. The two parts of the intron are not covalently linked, and thus generate a Yshaped molecule instead of a lariat. • The RNA that donates the 5' exon for transsplicing is called the SL RNA (spliced leader RNA) and exists as SLRNPs • The SL RNA can carry out the functions that the U1 snRNA performs at the 5’ splice site. • The trans-splicing reaction of the SL RNA may represent a step toward the evolution of the pre-mRNA splicing apparatus. tRNA SPLICING • The splicing of tRNA genes is achieved by a different mechanism that relies upon separate cleavage and ligation reactions. • The introns in tRNA genes representing different amino acids are unrelated. • There is no consensus sequence that could be recognized by the splicing enzymes. • All the introns include a sequence that is complementary to the anticodon of the tRNA.This creates an alternative conformation for the anticodon arm in which the anticodon is base paired to form an extension of the usual arm. • The exact sequence and size of the intron is not important. • Splicing oftRNA depends principally on recognition ofa common secondary structure in tRNA rather than a common sequence ofthe intron. • Regions in various parts of the molecule are important including the stretch between the acceptor arm and D arm, in the 1\If C arm, and especially the anticodon arm. • The two separate stages of the reaction are catalyzed by different enzymes. • • The first step does not require ATP. It involves phosphodiester bond cleavage by an atypical nuclease reaction. It is catalyzed by an endonuclease. • • The second step requires ATP and involves bond formation; it is a ligation reaction, and the responsible enzyme activity is described as an RNA ligase. • An endonuclease recognises introns and cleaves at both ends of the introns. SUMMARY • Splicing accomplishes the removal of introns and the joining of exons into the mature sequence of RNA. • The systems include eukaryotic nuclear introns, group I and group II introns, and tRNA introns. • Each reaction is usually a cis-acting event. • Consensus sequences • GU-AG rule • Transesterification and lariat formation • Spliceosome formation, Autosplicing , Alternative Splicing, Trans splicing , tRNA splicing REFERENCE • Benjamin Lewin,GENES IX , pg.667 – 695 • Watson,Baker et.al , Molecular Biology of the Gene,pg. 379 - 409