* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download CV - The Solomon H Snyder Department of Neuroscience
Development of the nervous system wikipedia , lookup
Neurotransmitter wikipedia , lookup
Axon guidance wikipedia , lookup
Activity-dependent plasticity wikipedia , lookup
Metastability in the brain wikipedia , lookup
Neuroeconomics wikipedia , lookup
Synaptic gating wikipedia , lookup
Nervous system network models wikipedia , lookup
Neurogenomics wikipedia , lookup
Endocannabinoid system wikipedia , lookup
Synaptogenesis wikipedia , lookup
Gene expression programming wikipedia , lookup
Molecular neuroscience wikipedia , lookup
Feature detection (nervous system) wikipedia , lookup
Signal transduction wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Neuroanatomy wikipedia , lookup
Channelrhodopsin wikipedia , lookup
Stimulus (physiology) wikipedia , lookup
Olfactory memory wikipedia , lookup
Optogenetics wikipedia , lookup
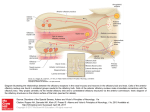
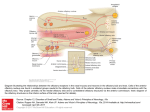
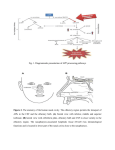
OMB No. 0925-0001 and 0925-0002 (Rev. 10/15 Approved Through 10/31/2018) BIOGRAPHICAL SKETCH Provide the following information for the Senior/key personnel and other significant contributors. Follow this format for each person. DO NOT EXCEED FIVE PAGES. NAME: Potter, Christopher John eRA COMMONS USER NAME (credential, e.g., agency login): cpotter4 POSITION TITLE: Assistant Professor, Department of Neuroscience EDUCATION/TRAINING (Begin with baccalaureate or other initial professional education, such as nursing, include postdoctoral training and residency training if applicable.) INSTITUTION AND LOCATION DEGREE Completion FIELD OF STUDY (if applicable) Date MM/YYYY University of California, Berkeley, Berkeley, BA 05/1996 Molecular Biology and California Genetics Yale University School of Medicine, New PHD 05/2002 Genetics Haven, CT Stanford University, Stanford, California Postdoctoral 03/2010 Neuroscience; Mentor: Liqun Fellow Luo A. Personal Statement The goal of my research is to use the vinegar fly, Drosophila melanogaster, as a genetically tractable insect model for characterizing the molecular and cellular components that direct innate odor guided repulsion or attraction. I am well suited for these studies. I have been conducting research with Drosophila for the past 20 years. As a graduate student at Yale, I used genetic screens to identify and characterize novel mutations in a signal transduction pathway, and performed biochemical analyses on the gene products. As a Postdoctoral Fellow at Stanford, I helped develop and implement an approach known as brain warping whereby we generated hundreds of singly labeled neurons in the fly brain and used computer software to align hundreds of different confocal images with one another. I then utilized computer algorithms to model how these neurons organize a part of the brain. I have a track record of developing new genetic reagents in Drosophila for investigating biological functions. As a Postdoc, I developed a new repressible binary expression system (Qsystem) that I used for generating intersectional expression patterns to limit analyses to small neuronal populations. In collaboration with the Clandinin lab, I was involved in the development of the inSITE system that allows genes of interest, by a series of crosses, to be swapped into previously genomically integrated transgenes. In collaboration with the Luo lab, I was involved in the development of a transcriptional reporter for neuron activity (TRIC). In collaboration with the White lab, I was involved in the development of an artificial expression exon for capturing gene expression patterns (MiMIC-TIFF). My lab has also recently developed a novel genetic approach called CLAMP (Cell Labeling Across Membrane Partners) that allows us for the first time to identify neurons downstream of olfactory receptor and projection neurons based on connectivity. This approach is not limited to olfactory neurons however. It is based on an intersectional approach that labels cells within close proximity. We have developed two versions of this method to label cells in close contact- one which uses secretion of a signal and the second that uses synaptic localization of a signal. The secretion method is best suited to identify the connectivity of neurosecretory or glial cells, and the synaptic method is best suited for analyzing synaptic connectivity. I have recruited students and Postdocs, and have created an exciting research atmosphere. In sum, I have the environment, experience, and the know-how to accomplish my research goals. 1. Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, Potter CJ. Improved and expanded Q-system reagents for genetic manipulations. Nat Methods. 2015 Mar;12(3):219-22, 5 p following 222. PubMed PMID: 25581800; PubMed Central PMCID: PMC4344399. 2. Gao XJ, Potter CJ, Gohl DM, Silies M, Katsov AY, Clandinin TR, Luo L. Specific kinematics and motorrelated neurons for aversive chemotaxis in Drosophila. Curr Biol. 2013 Jul 8;23(13):1163-72. PubMed PMID: 23770185; PubMed Central PMCID: PMC3762259. 3. Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, Lin CC, Potter CJ, Clandinin TR. A versatile in vivo system for directed dissection of gene expression patterns. Nat Methods. 2011 Mar;8(3):231-7. PubMed PMID: 21473015; PubMed Central PMCID: PMC3079545. 4. Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010 Apr 30;141(3):536-48. PubMed PMID: 20434990; PubMed Central PMCID: PMC2883883. B. Positions and Honors Positions and Employment 2002 - 2010 2010 - Postdoctoral Fellow, Stanford University, Stanford, CA Assistant Professor, Johns Hopkins University School of Medicine, Baltimore, MD Other Experience and Professional Memberships 2010 2014 - Member, Society for Neuroscience Member, Association for Chemoreception Sciences Honors 1996 2003 2011 Outstanding Undergraduate Geneticist Award, UC Berkeley Damon Runyon Cancer Research Fellowship, Stanford University Whitehall Foundation Fellowship, Johns Hopkins University School of Medicine C. Contribution to Science 1. Scientific progress often follows the invention of new experimental methods. The adoption of the GAL4/UAS system revolutionized experimental research in Drosophila. Yet despite its versatility, the GAL4 system had limitations. For example, GAL4 expression patterns from enhancer trap lines or promoterdriven transgenes often include cells besides the cells of interest. It is thus difficult to assign the effect of transgene expression to a specific cell population, especially when phenotypes, such as behavior, are assayed at the whole organism level. Additionally, analysis of gene function and dissection of complex biological systems in multicellular organisms often requires independent genetic manipulations of separate populations of cells. This is not possible with a single binary expression system. We developed the repressible binary QF/QUAS system to address these limitations in experimental research. In a single tour-de-force paper, we introduced Q-system applications that had taken multiple labs over 20 years to develop for the GAL4 system (e.g., transgenic animals, enhancer traps, tissue specific promoter expression, introduction of a transcription factor suppressor, mosaic analyses, MARCM analyses, behavioral analyses). In addition, unlike the GAL4 system, we also demonstrated that the Q-system could be regulated by feeding animals a non-toxic drug. We further introduced and validated new intersectional genetic approaches for refining existing GAL4 expression patterns. These genetic approaches are essential for most current experimental studies, and allowed us to assign olfactory functions to a small neuronal population. The Q-system is a versatile platform for experimental innovation and discovery by researchers worldwide. a. Subedi A, Macurak M, Gee ST, Monge E, Goll MG, Potter CJ, Parsons MJ, Halpern ME. Adoption of the Q transcriptional regulatory system for zebrafish transgenesis. Methods. 2014 Apr 1;66(3):433-40. PubMed PMID: 23792917; PubMed Central PMCID: PMC3883888. b. Wei X, Potter CJ, Luo L, Shen K. Controlling gene expression with the Q repressible binary expression system in Caenorhabditis elegans. Nat Methods. 2012 Mar 11;9(4):391-5. PubMed PMID: 22406855; PubMed Central PMCID: PMC3846601. c. Potter CJ, Luo L. Using the Q system in Drosophila melanogaster. Nat Protoc. 2011 Jul 7;6(8):110520. PubMed PMID: 21738124; PubMed Central PMCID: PMC3982322. d. Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010 Apr 30;141(3):536-48. PubMed PMID: 20434990; PubMed Central PMCID: PMC2883883. 2. Although immediately useful, the QF transcription factor we described previously had a problem: we could not generate pan-neuronal or pan-tissue transgenic animals expressing QF. This suggested that QF, if expressed broadly, caused lethality. The reason for this was unknown. This publication described the solution to this problem. To determine which region of QF was the cause of lethality, we decided to generate chimeric QF proteins that swapped QF domains with GAL4 domains. We separated the QF protein into 3 domains: the DNA binding domain, the middle domain, and the transactivator activation domain. GAL4 was divided similarly. We generated constructs for all possible chimeric combinations between QF and GAL4 for expression in vitro cell culture activity assays, and for generating pan-neuronal transcriptional activator lines. To our surprise, we identified that any construct that contained the QF middle domain led to toxicity. Even more surprising, the middle domain was not necessary for in vivo function: QF variants containing only the QF DNA binding domain and activation domain were fully functional, and led to healthy transgenic animals. We generated pan-neuronal and pan-tissue lines using this new variant, which we named QF2. We further showed, via extensive behavioral analyses, that pan-neuronal QF2 expression had no effect on neuronal development or function. These new lines will have enormous value in developmental and behavioral studies. Most importantly, QF2 allows expansion of the Q-system into other animals, such as mice, zebrafish, and plants, which might have presented lethality with the original QF. We have introduced QF2 into the mosquito system, and it has proven to be effective and healthy. In addition to QF2, we also generated functional chimeras between GAL4, QF, and LexA. These can be used for novel genetic intersectional experiments and greatly expanded the flexibility and power of transgenic manipulations a. Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, Potter CJ. Improved and expanded Q-system reagents for genetic manipulations. Nat Methods. 2015 Mar;12(3):219-22, 5 p following 222. PubMed PMID: 25581800; PubMed Central PMCID: PMC4344399. 3. Besides the methods described above, I collaborate with many groups that share an interest in developing new genetic reagents. I have primarily contributed to introducing Q-system reagents into these new genetic toolkits, but also aid in experimental design and approaches. a. Gao XJ, Riabinina O, Li J, Potter CJ, Clandinin TR, Luo L. A transcriptional reporter of intracellular Ca(2+) in Drosophila. Nat Neurosci. 2015 Jun;18(6):917-25. PubMed PMID: 25961791; PubMed Central PMCID: PMC4446202. b. Diao F, Ironfield H, Luan H, Diao F, Shropshire WC, Ewer J, Marr E, Potter CJ, Landgraf M, White BH. Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell Rep. 2015 Mar 3;10(8):1410-21. PubMed PMID: 25732830; PubMed Central PMCID: PMC4373654. c. Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, Lin CC, Potter CJ, Clandinin TR. A versatile in vivo system for directed dissection of gene expression patterns. Nat Methods. 2011 Mar;8(3):231-7. PubMed PMID: 21473015; PubMed Central PMCID: PMC3079545. 4. We developed a practical and useful update to a commonly used method for investigating olfactory signaling. Drosophila olfactory organs are covered in specialized sensory hairs called sensilla. A powerful technique for investigating olfactory neuron activities in these hairs is called Single Sensillum Recordings (SSR), whereby an electrode is placed within a sensilla to record the activities of the neurons within the sensilla. In this manner, the activities of the neurons can also be monitored upon stimulation with odorants. There are 22 different types of sensilla containing olfactory receptor neurons in Drosophila, of differing abundances, and identifying which sensilla is being recorded from usually requires careful comparison of odor response profiles to known standards. While powerful, SSR can be tedious, time consuming, and subject to error, especially when attempting to systematically screen all known sensilla. There have been no major improvements to this technique in 30 years. We combined SSR with the Gal4/UAS system to allow specific sensilla, based on the olfactory neurons found within that sensilla, to be targeted for recordings. This greatly increased the speed and accuracy of the SSR method. We validated this method, termed Fluorescence-guided SSR (FgSSR), for use in targeting any olfactory receptor neuron. In this manner, a systematic approach can now be used to screen specific sensilla, or to target all sensilla. We provide the necessary information and reagents to allow easy adoption of this technique by other researchers. Using FgSSR, we determined that two types of sensilla, the trichoids and intermediate, have been classified incorrectly. We presented detailed analyses of these sensilla, and presented a re-classification based on our data. This reclassification is a necessary addition to the literature, as it prevents confusion and will make future studies on intermediate and trichoid sensilla more easily comparable. a. Lin CC, Potter CJ. Re-Classification of Drosophila melanogaster Trichoid and Intermediate Sensilla Using Fluorescence-Guided Single Sensillum Recording. PLoS One. 2015;10(10):e0139675. PubMed PMID: 26431203; PubMed Central PMCID: PMC4592000. 5. We characterized a previously unreported pheromone-signaling pathway in Drosophila. By utilizing a large arena four-field olfactory behavioral assay, we discovered that male flies, when stimulated by food-odors, deposit a pheromone that influences many behaviors associated with food sites, such as aggregation and egg-laying site selection. To our knowledge, this is the first cuticular male pheromone identified that influences female egg-laying decisions. By GC-MS, we identified this pheromone as 9-tricosene. Using behavioral, genetic, and electrophysiological methods, we determined the molecular and neuronal mechanisms by which 9-tricosene functions to alter Drosophila behaviors. Specifically, we identified the odorant receptor Or7a as being necessary and sufficient for mediating pheromone behaviors. This work introduced the concept that food-odor-perception can guide pheromone signaling, identified a pheromone responsible for regulating egg-laying behaviors of the opposite sex, pinpoints the molecular receptor that detects the pheromone, and demonstrates that this receptor is necessary and sufficient for the observed behaviors. The work also introduced the following new concepts and advancements: 1. We identified the olfactory receptor neuron antennal basiconic 4A (ab4A) as the most strongly activated by the pheromone 9-tricosene. This was surprising as pheromone signaling in Drosophila was previously considered specific to olfactory neurons housed in trichoid sensilla. This is the first example, to our knowledge, of a Drosophila pheromone that activates non-trichoid olfactory receptor neurons, and suggests that basiconic sensilla are responsive to a greater range of stimuli than previously appreciated. 2. The ab4A olfactory neuron expresses the Or7a olfactory receptor. By ectopically expressing Or7a in olfactory neurons that lack their endogenous odorant receptors, we found that Or7a is both necessary and sufficient for 9-tricosene stimulated activities. This represents the first cuticular hydrocarbon pheromone to be matched to an odorant receptor. These studies established a new model for characterizing pheromone signaling in a genetic system. 3. The Or7a olfactory receptor can also be activated by the green leaf volatile E2-hexenal. We found that this odorant, like 9-tricosene, can guide aggregation and egg-laying decisions. This suggests that odorant-stimulated activity of Or7a receptors may play a key role in female oviposition. 4. Previous studies suggested the pheromone cVA as being the predominant Drosophila aggregation pheromone. We present evidence that 9-tricosone is a major Drosophila aggregation pheromone in response to food odors. 5. Our data suggests that attractive odors per se are not sufficient to trigger pheromone deposition, but instead it is the perception of food-odors that drives pheromone deposition. To our knowledge, this is the first reported case in which a) a pheromone is rapidly deposited upon an odor-stimulation, and b) an olfactory perception (food odor) has been reproduced by a mixture of known constituent components. This will prove critical in future studies aimed at understanding how sensory perceptions are encoded in the brain. 6. The Or7a receptor is also activated by a number of other odorants besides 9-tricosene, include many alchohols and aldehydes. We found that the specificity of the Or7a response directly correlated to an egg-laying decision. Odorants, like 9-tricosene, that most specifically activated Or7a led to the greatest oviposition preference. Other odorants, like benzaldehyde, which activate Or7a but many other Odorant receptors, led to weaker oviposition preference. This highlights how an animal can use an odorant receptor that responds to many different odorants to guide a specific olfactory behavior. This also likely reflects how the olfactory system is utilized to make decisions based on complex odor mixtures. a. Lin CC, Prokop-Prigge KA, Preti G, Potter CJ. Food odors trigger Drosophila males to deposit a pheromone that guides aggregation and female oviposition decisions. Elife. 2015 Sep 30;4PubMed PMID: 26422512; PubMed Central PMCID: PMC4621432. D. Additional Information: Research Support and/or Scholastic Performance Ongoing Research Support R01 DC013079-01 Potter, Christopher John (PI) 03/25/13-02/28/18 Neural circuits mediating food, pherome, and repulsive odor behaviors Role: PI R21 NS088521-01A1 Wu, Mark N (Co-PI) 02/01/15-01/31/17 Developing Genetic Reagents for the Dissection of Dopaminergic Circuitry Role: Co-PI MRI-Potter_2015, Malaria Research Institute at Johns Hopkins Potter, Christopher (PI) 07/01/15-06/30/16 Genetic dissection and characterization of the Anopheles gambiae olfactory system Role: PI Completed Research Support Synergy_Potter2014, Johns Hopkins University School of Medicine Potter, Christopher (PI) 07/01/14-06/30/15 Imaging sensory activity in the Anopheles gambiae mosquito brain. Role: CPI 2011-05-04, Whitehall Foundation Potter, Christopher (PI) 07/01/11-06/30/14 Neuronal Dissection of Olfactory Behaviors in Drosophila. Role: PI