* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download SEGMENTAL VARIATION
Cre-Lox recombination wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene expression profiling wikipedia , lookup
Y chromosome wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Gene desert wikipedia , lookup
Minimal genome wikipedia , lookup
Epigenomics wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Transposable element wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
History of genetic engineering wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Segmental Duplication on the Human Y Chromosome wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Neocentromere wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Public health genomics wikipedia , lookup
X-inactivation wikipedia , lookup
Designer baby wikipedia , lookup
Non-coding DNA wikipedia , lookup
Genome (book) wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Frameshift mutation wikipedia , lookup
DNA sequencing wikipedia , lookup
Oncogenomics wikipedia , lookup
Copy-number variation wikipedia , lookup
Human genome wikipedia , lookup
Microevolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Pathogenomics wikipedia , lookup
Genome evolution wikipedia , lookup
Point mutation wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Helitron (biology) wikipedia , lookup
Human Genome Project wikipedia , lookup
Genomic library wikipedia , lookup
Genome editing wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
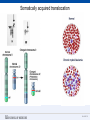
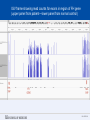
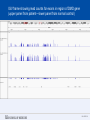
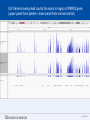
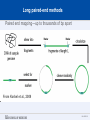
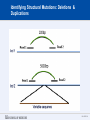
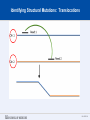
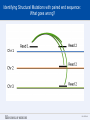
SEGMENTAL VARIATION (Copy Number Variants and other gross chromosomal rearrangements) Allen E. Bale, M.D. Dept. of Genetics SLIDE 0 Importance of Copy Number Variants (CNVs) and Other Rearrangements in Health and Disease • Constitutional (germ-line) variants in hereditary conditions – Large and small copy number variants – Translocations and inversions: rarely cause a phenotype but may generate CNVs due to mis-pairing during meiosis • Somatically acquired variants in cancer – Duplications and deletions: amplification of oncogene; loss of tumor suppressor – Translocations and inversions: place oncogene under control of an active promoter SLIDE 1 What is the origin of structural variants? • An area of active research • Recurrent constitutional CNVs: Often related to illegitimate recombination between homologous, but non-identical, sequences • Rare, non-recurrent, constitutional CNVs: No obvious sequence homology at breakpoints, ?non-homologous end joining • Tumor CNVs: Any mechanism to create a rearrangement that favors tumor growth, often non-homologous end joining. SLIDE 2 Cytogenetically visible CNVs and translocations SLIDE 3 A Really Large CNV SLIDE 4 Somatically acquired translocation SLIDE 5 Limitations of Cytogenetics • Cell has to be proliferating in order to arrest chromosomes at metaphase (when they are visible under the microscope) • Resolution is limited (in the range of 5 Mb) • Requires highly skilled technologists and still a lot of handson time, even with sophisticated image processing SLIDE 6 Submicroscopic CNVs: Array CGH* *Frequently referred to as “chromosome microarray” SLIDE 7 Example: Submicroscopic 22q deletion • Abnormal nose, ears, and palate • Also heart, parathyroid, and thymus abnormalities SLIDE 8 Limitations of Array CGH • Can’t detect translocations and inversions • Resolution still limited by number of probes on the array—typical resolution about 100 kb • Still a fair amount of variability in results depending on exactly which array is used SLIDE 9 Genome-scale sequencing to detect rearrangements If you could sequence each chromosome as one continuous piece of DNA, from one end to the other with no gaps in the sequence, what structural variants would you miss? S L I D E 10 Genome-scale sequencing to detect rearrangements What methods are currently in use? •Depth-of-coverage methods Regions that are deleted or duplicated should yield lesser or greater numbers of reads •Detection of breakpoints by: –Short paired reads (like Illumina paired-end sequencing) Are the sequences at two ends of a fragment both from the same chromosome? Are they the right distance apart? –Long reads (kb-scale) Direct sequencing of breakpoints S L I D E 11 Genome-scale sequencing to detect rearrangements •Depth-of-coverage method •Detection of breakpoints by short paired reads •Detection of breakpoints by long reads Compared with cytogenetics and array CGH, how would the approaches above perform? • What would be missed by depth-of-coverage reading? • What would be missed by detection of breakpoints? • What problems do you foresee with these two approaches? S L I D E 12 Depth-of-coverage example: Whole exome sequencing as a tool to identify both sequence variants and CNVs S L I D E 13 Whole exome sequencing • Capture portions of the genome containing exons in order to efficiently sequence coding regions • Not designed for CNV detection, but potentially contains information on gene dosage • For any gene, the number of fragments captured on the array and sequenced should be proportional to the representation in the starting material S L I D E 14 Array CGH vs. Exome Sequencing S L I D E 15 Does this work at all? • Total reads on the X chromosome were counted in a series of males and females • Gene dosage for the X chromosome in males should be half the gene dosage for the X chromosome in females S L I D E 16 Does it work for single exons? Reads counted for each exon of the OTC gene on X chromosome Males should have one half the female dosage. • Read number varies among exons due to different capture efficiencies but is consistent subject to subject. • Exons with sufficient read numbers show dosage effect. • Performs very well for this 70 kb gene taken as a single unit. S L I D E 17 Approach to scanning the whole genome for CNVs • The genome was divided into 50 kb windows. • Intervals with zero reads were removed. • Mean number of reads and standard deviations for each interval were calculated from 10 exome sequences. • Depth of coverage in a single patient was compared to average and standard deviation of depth of coverage. • Algorithms were developed for: – Classifying X chromosome as being deleted in males compared with females – Classifying X chromosome as being duplicated in females compared with males S L I D E 18 Chromosomal coverage with non-zero, 50 kb intervals corresponds exactly to density of coding sequences S L I D E 19 Test case: Female with a 338 kb duplication on 5q35 Diagram shows all loci passing initial algorithm S L I D E 20 Filter #1: Require two adjacent intervals to both be deleted or duplicated S L I D E 21 Filter #2: Remove “deleted regions” that contain heterozygous variants S L I D E 22 Filter # 3: Remove intervals with read counts <200 S L I D E 23 Application to 7 subjects with deletions or duplications in 500 kb to 1 Mb range S L I D E 24 Another reduced representation method: Alu-PCR* Mei et al. BMC Genomics 2011, 12:564 S L I D E 25 Some problems with use of reduced representation data • Intervals with no genes are not covered in exome (important?) • Intervals having close homologs elsewhere in the genome can not be accurately evaluated. • Because this technology is evolving rapidly, the normal standard to which a test sample is compared needs to be a pool of recent sequences generated by identical methods (huge FDR with nonhomogeneous samples). S L I D E 26 What works well: Targeted analysis of candidate genes Case 1 • 30 year old woman with personal and family history of renal cell cancer • Scanning candidate genes for point mutations or small indels: o o o o o o o o o o SDHB SDHC SDHD FH VHL MITF BAP1 MET FLCN HNF1B Neg Neg Neg Neg Neg Neg Neg Neg Neg Neg S L I D E 27 IGV frame showing read counts for exons in region of FH gene (upper panel from patient—lower panel from normal control) S L I D E 28 Case 2 • One year old male with low muscle tone and strength and poor cardiac function • No family history of a similar disease • Scanning entire exome for point mutations or small indels: • No mutations found S L I D E 29 IGV frame showing read counts for exons in region of DMD gene (upper panel from patient—lower panel from normal control) S L I D E 30 Case 3 • 25 year old woman with numbness and weakness of lower legs • Similar or more severe features in several relatives • Scanning of entire exome for point mutations and small deletions: • Negative S L I D E 31 IGV frame showing read counts for exons in region of PMP22 gene (upper panel from patient—lower panel from normal control) S L I D E 32 For a review of published depth-of-coverage methods for exome or genome data see: Klambauer, G. et. al. (2012). "cn.MOPS: mixture of Poissons for discovering copy number variations in nextgeneration sequencing data with a low false discovery rate." Nucleic Acids Res. Compares several programs, none of which work really well. Two newer programs for exome sequencing are in your reading list. S L I D E 33 Paired-end methods • Illumina HiSeq, the current industry leader in highthroughput sequencing, generates short reads from fragments 200 to 600 bp long. • Reading both ends of the same fragment gives you sequences that should lie 200 to 600 bp apart • Other methods can generate paired fragments that lie even farther apart S L I D E 34 Long paired-end methods Paired end mapping—up to thousands of bp apart From Korbel et al., 2009 S L I D E 35 Identifying Structural Mutations: Deletions & Duplications S L I D E 36 Identifying Structural Mutations: Inversions S L I D E 37 Identifying Structural Mutations: Translocations S L I D E 38 Analyzing structural variations from paired end data • PEMer (Korbel et al., 2009): For discovery of CNVs and inversions; could also be implemented for translocations • Breakdancer (Chen et al., 2009): For discovery of CNVs, inversions, and translocations S L I D E 39 Identifying Structural Mutations with paired end sequence: What goes wrong? S L I D E 40 How to overcome problems with paired end detection of CNVs Separating the wheat from the chaff • Technical artifacts (ligation of unrelated fragments during library preparation) may be numerous but will be random • Artifacts related to homologous sequences (see previous slide) will be reproducible but common to all samples • Real structural variants will be reproducible within a sample and not common to all samples • How much reading depth do you need to detect the real variants? S L I D E 41 Toward direct sequencing of breakpoints • Long reads – PACbio can generate reads of 1000 bp or so – Nanopore sequencing said to generate reads in the 10s of thousands • Strobe sequencing with PACbio: Normally read length is limited due to inactivation of polymerase by laser. Short bursts of laser give sample sequences along a stretch of DNA in the 20 kb range. S L I D E 42 The poor man’s strobe sequencing: 10X Genomics Isolate a very long, single DNA fragment and tag it with a molecular barcode at many locations along it’s length: Molecular barcode DNA molecule 100 kb long Fragment the large DNA molecule into 250 bp pieces and sequence using Illumina technology. Use informatics to link the short reads into a long haplotype. S L I D E 43 10X Genomics technology: Parallel barcoding of many long fragments Zheng GXY et al., Nature Biotechnology 34:303, 2016 S L I D E 44 S L I D E 45 Programs for analysis of longer reads that directly sequence breakpoints • CREST (Wang et. al., 2011): Detects small and large structural variants by direct sequencing of breakpoints. • SRiC (Zhang et al., 2011): Similar to CREST • Algorithm for strobe reads (Ritz et al., 2010) • Several methods reviewed in Abel and Duncavage, 2014 S L I D E 46 Conclusions • Structural variation in the genome accounts for a great deal of human phenotypic variability including disease • Depth-of-coverage methods can detect many CNVs but not inversions and translocations. Variation from sample to sample limits sensitivity and specificity. • Whole genome sequencing, which can identify all types of structural variants, will supersede depth-of-coverage methods. • Large scale and small scale duplications and repetitive sequences remain a major obstacle. S L I D E 47