* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Cell and Molecular Biology
Expression vector wikipedia , lookup
Biochemistry wikipedia , lookup
Gene regulatory network wikipedia , lookup
Alternative splicing wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Western blot wikipedia , lookup
Non-coding DNA wikipedia , lookup
Point mutation wikipedia , lookup
RNA silencing wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Paracrine signalling wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Metalloprotein wikipedia , lookup
Genetic code wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Transcription factor wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Proteolysis wikipedia , lookup
Messenger RNA wikipedia , lookup
Biosynthesis wikipedia , lookup
Polyadenylation wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Gene expression wikipedia , lookup
Epitranscriptome wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
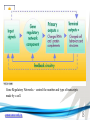
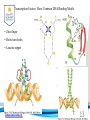
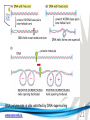
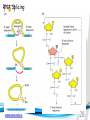
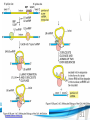
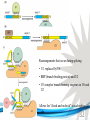
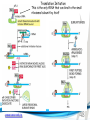
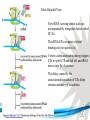
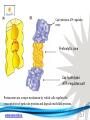
Cell and Molecular Biology DNA Transcription Behrouz Mahmoudi www.soran.edu.iq 1 1. What are two major differences between transcription in prokaryotic and eukaryotic cells? 2. RNA polymerase and DNA polymerase enzymes catalyze the “same” reaction, but there are some distinct differences in what is required to make them begin catalysis and end catalysis. What are these differences? 3. Which is more accurate, DNA replication or RNA transcription? 4. Explain the proteins and mechanisms involved in the initiation of transcription 5. What determines how many copies of a transcript (mRNA) are made? 6. How are elongation and termination of the transcript regulated? www.soran.edu.iq 2 “The Protein Players” - RNA polymerases, transcription factors, initiation factors, enhancers, repressors www.soran.edu.iq 3 www.soran.edu.iq 4 Prokaryotic Transcription www.soran.edu.iq 5 DNA Sequences Important to Transcription Prokaryotes • Promoter – •Pribnow Box (also called the -10 element) – TATAAT •-35 element - TTGACA Eukaryotes • Promoter –(asymmetrical sequence) • Basic core promoter –TATA box (TATAAA(A)); within 50bp upstream of start site; found in unicellular eukaryotes • Core promoter PLUS •Downstream promoters •Proximal promoter elements www.soran.edu.iq 6 • Initiator sequences • Regulatory Elements/Response Element - Response elements are the recognition sites of certain transcription factors Most of them are located within 1 kb from the transcriptional start site. • Enhancer elements -upon binding with transcription factors (activators), can enhance transcription; located either upstream or downstream of the transcriptional initiation site. •Upstream enhancer elements •Downstream enhancers •Distal enhancer elements • Silencers - upon binding with transcription factors (repressors), can repress transcription. • Insulators www.soran.edu.iq 7 Gene Regulatory Networks – control the number and type of transcripts made by a cell. www.soran.edu.iq 8 www.soran.edu.iq 9 Simple core promoter UAS = upstream activator sequence RE = regulatory elements www.soran.edu.iq downstream promoter elements INR = initiator sequence 10 DPE = Sequences that can act as promoters (TATA is preferred) www.soran.edu.iq 11 Proteins Involved in Transcription RNA Polymerase General (or Basal) Transcription Factors: TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH Transcription Factors that Bind to Regulatory Elements Holoenzyme or Initiation Complex www.soran.edu.iq 12 Transcription Factors Have Common DNA Binding Motifs • Zinc finger • Helix-turn-helix • Leucine zipper www.soran.edu.iq 13 RNA Polymerase www.soran.edu.iq 14 www.soran.edu.iq 15 Recognizes and binds to TATA box; TBP + 10 TBP associated factors; position set www.soran.edu.iq 16 TBP bends DNA ~80o and forces open the minor groove. www.soran.edu.iq 17 Recognizes and binds to TATA box; TBP + 10 TBP associated factors Binds and stabilizes the TFIID complex Recruits RNA pol II + TFIIF to the location Two subunits - RAP38 & RAP74. Rap74 has a helicase activity; RAP38 binds RNAPolII Two subunits - recruits TFIIH to the complex thereby priming the initiation complex for promoter clearance and elongation complex of 9 subunits. One w/ kinase activity; one w/ helicase activity; one is a cyclin (cdk7) www.soran.edu.iq 18 -30 +1 TATA Inr TAFs Sequential Binding Model for assembly of preinitiation complex } TBP TFIID or TBP IIA IIB Eukaryotic RNA polymerase II IIF Pol IIa CTD of large subunit of Pol II IIE helicase IIH protein kinase IIF IIE IIA IIB TATA Inr Pol IIa IIH preinitiation complex = PIC ATP hydrolysis IIE Polymerization of 1st few NTPs and phosphorylation of CTD leads to promoter clearance. TFIIB, TFIIE and TFIIH dissociate, PolII+IIF elongates, andwww.soran.edu.iq TFIID + TFIIA stays at TATA. IIA IIB TATA IIF Pol IIa IIH Inr initiation complex, DNA melted at Inr Activated PIC 19 Transcription initiation in the cell often requires the local recruitment of chromatinmodifying enzymes, including chromatin remodeling complexes and histone acetylases greater accessibility to the DNA present in chromatin www.soran.edu.iq 20 RNA polymerase is also assisted by DNA supercoiling www.soran.edu.iq 21 Phosphorylation of the carboxy terminal domain (CTD) of one of the subunits of RNA PolII; RNA polymerase II dissociates from the transcription factors and other protein complexes that were required for assembly and elongation begins Phosphorylation also promotes the accumulation of elongation factors – other proteins that arrest transcription long enough to recruiting RNA processing enzymes www.soran.edu.iq 22 Elongation is Coupled to RNA Processing • Capping • Splicing • Polyadenylation www.soran.edu.iq 23 www.soran.edu.iq 24 www.soran.edu.iq 25 RNA Capping enzymes: • Phosphatase • Guanyl transferase – GMP in 5’ to5’ linkage • methyltransferase www.soran.edu.iq Video of transcription and capping 26 CBC – cap binding complex proteins also associate and protect the cap; Later they will direct transcript in its exit from the nucleus www.soran.edu.iq 27 RNA Splicing www.soran.edu.iq 28 How Introns Are Identified: Consensus sequences at (5’ to 3’ direction) •5’ splice site •Lariate loop closure site of the intron sequence •3’ splice site www.soran.edu.iq R=A or G,Y=C or U 29 The Spliceosome Forms snRNAs (U1, U2, U4, U5 and U6) and associated proteins = snRNPs • U1 binds to the GU sequence at the 5' splice site, along with accessory proteins/enzymes, • U2 binds to the branch site, and ATP is hydrolyzed; • U5/U4/U6 trimer binds, and the U5 binds exons at the 5' site, with U6 binding to U2; • U1 is released, U5 shifts from exon to intron and the U6 binds at the 5' splice site; • U4 is released, U6/U2 catalyzes transesterification, U5 binds exon at 3' splice site, and the 5' site is cleaved, resulting in the formation of the lariat; • U2/U5/U6 remain bound to the lariat, and the 3' site is cleaved and exons are ligated using ATP hydrolysis. The spliced RNA is released and www.soran.edu.iq the lariat debranches. 30 www.soran.edu.iq 31 Rearrangements that occur during splicing • U1 replaced by U6 • BBP (branch binding protein) and U2 • U5 complex branch forming enzymes in U6 and U2 www.soran.edu.iq Allows for “check and recheck” at each splice site. 32 Why is splicing so accurate? Introns are small-large; Exons are about 150bp long Exons might be easily identified, while introns probably couldn’t be. www.soran.edu.iq 33 As the RNA is being transcribed, SR proteins (rich in serine (S) and arginine (R)) sit down on the exons. Along with the U proteins, demarcates the start and end of the exon. Capping proteins or polyA binding proteins act as markers at either end of the transcript. Other hnRNPs (heterogeneous nuclear RNPs) bind along the introns, helping to distinguish these sequences from exons. www.soran.edu.iq 34 Changes in splicing patterns caused by random mutations have been an important pathway in the evolution of genes. www.soran.edu.iq 35 3’ end splicing sequence • AAUAAA • Cleavage site CA – 10-30 nucleotides downstream • Polyadenylation site – GU- or U-rich region about 30 nucleotides downstream from the cleavage site www.soran.edu.iq 36 CPSF = Cleavage and Polyadenylation Specificity Factor CstF = Cleavage Stimulation Factor www.soran.edu.iq 37 Poly A polymerase – no template strand required All of the A nucleotides are derived from ATP Poly A binding proteins remain until mRNA undergoes translation www.soran.edu.iq 38 Only very “select” RNAs can be transported out of the nucleus www.soran.edu.iq 39 Guided diffusion along the FG-repeats displayed by nucleoporins Proteins bound to mature mRNA molecules and that signal completed splicing have nuclear export signals as a part of their sequence www.soran.edu.iq 40 •hNRPs “straighten out” the mature mRNA so that nuclear export signals can be “read” •5’ cap enters the pore first •Many of the RNA binding proteins fall off as mRNA exits the nucleus •Initiation factors (elF-4G and elF-4E) immediately bind to the 5’ capping complex (which falls off) and to the polyA tail, forming a loop www.soran.edu.iq 41 Test Your Knowledge – Translation • Transcription requires only changing a DNA code of nucleotides into a similar RNA code of nucleotides, while translation involves changing the RNA code into what? • What are codons and what “reads” codons? • What is “wobble” and how is it related to translation? • What attaches amino acids to t-RNA? • What are the “parts” of the ribosome? What function does each part perform? • What are the A, P, and E sites of a ribosome? What binds at each of these sites? • Does anything beside the ribosome participate in elongation of the amino acid chain? If so, what is it and what does it do? • What signals where translation starts and stops? • What happens to improperly translated or proteins that don’t fold properly after being translated? www.soran.edu.iq 42 Transfer RNA • anticodon- 3’ to 5’ sequence that matches the complementary 5’ to 3’sequence (codon) on the mRNA • Acceptor arm - Amino acid code on 3’ end • T and D loops – provide structure for interface with aminoacyl-tRNA synthetase ? www.soran.edu.iq 43 www.soran.edu.iq 44 A different aminoacyl-tRNA synthetase enzyme for each amino acid www.soran.edu.iq 45 www.soran.edu.iq 46 Large subunit does???? www.soran.edu.iq Small subunit does ????? 47 Translation Initiation This is the only tRNA that can bind to the small ribosomal subunit by itself www.soran.edu.iq 48 Protein made in 5’ to 3’ direction, with N-terminal end made first General Mechanism • A site is where new codon is translated • P site is where the growing peptide chain is kept and new aa are attached • E site is where “naked” t RNA exit the ribosome www.soran.edu.iq 49 More Detailed View New tRNA carrying amino acids are accompanied by elongation factor called EF-Tu The tRNA-ETu occupies a hybrid binding site (not quite in A) Correct codon-anticodon pairing triggers ETu to split GTP and fall off, and tRNA moves into the A position The delay caused by the association/dissociation of ETu helps increase accuracy of translation www.soran.edu.iq 50 Elongation factor G (EF-G) then binds near the A site, forcing the tRNAs containing the new amino acid and the growing chain into the next (P and E) sites on the ribosome EF-G splits GTP, changes conformation and falls off, thus increasing the speed of translation. GTP exchange factors continually recharge the GTP on both of the elongation factors. www.soran.edu.iq 51 Stop Codons = UAA, UAG, UGA No tRNA binds to this set of codons One of these codons at the A site attracts a release factor Ribosome adds a water to the last peptide, creating the carboxyl end www.soran.edu.iq 52 www.soran.edu.iq 53 Proteins Fold as They are Translated on the Ribosome www.soran.edu.iq 54 Hsp 70 works to help fold early in the lifecycle of a protein Hsp 60 works like a quality control chamber after a protein is completely folded www.soran.edu.iq 55 www.soran.edu.iq 56 Cap hydrolyses ATP; regulates entry Proteolytic core Cap hydrolyses ATP; regulates exit Proteasomes are a major mechanism by which cells regulate the concentration of particular proteins and degrade misfolded proteins. www.soran.edu.iq 57 Proteins are marked for destruction by the addition of a small molecule called ubiquitin on exposed lysine residues www.soran.edu.iq 58 www.soran.edu.iq 59 Cells can also regulate protein degradation by activating new ubiquitin ligases via different mechanisms. www.soran.edu.iq 60 Proteins generally “hide” degradation signals so that they are not active. However, there are several mechanisms for exposing the degradation signals. www.soran.edu.iq 61 www.soran.edu.iq 62 You have isolated an antibiotic named edeine, from a bacterial culture. Edeine inhibits protein synthesis but has not effect on either DNA synthesis or RNA synthesis. When added to a reticulocyte lysate, edeine stops protein synthesis after a short lag, as shown below. By contrast, cycloheximide stops protein synthesis immediately. Analysis of the edeineinhibited lysate by density-gradient centrifugation showed that no polyribosomes remained by the time protein synthesis had stopped. Instead, all the globin mRNA accumulated in an abnormal 40S peak, which contained equimolar amounts of the small ribosomal subunit and initiator tRNA. A.What step in protein synthesis does edeine inhibit? B.Why is there a lag between addition of edeine and cessation of protein synthesis? What determines that lag? Radioactivity in hemoglobin C.Would you expect the polyribosomes to disappear if you added cycloheximide at the same time as edeine? www.soran.edu.iq control Add inhibitor edeine cycloheximide 0 2 4 Time (min) 6 8 63