* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download myPresentation
Metabolic network modelling wikipedia , lookup
Transposable element wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene desert wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Quantitative trait locus wikipedia , lookup
RNA silencing wikipedia , lookup
Essential gene wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Metagenomics wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Pathogenomics wikipedia , lookup
Gene expression programming wikipedia , lookup
History of genetic engineering wikipedia , lookup
RNA interference wikipedia , lookup
Public health genomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome evolution wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Microevolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Designer baby wikipedia , lookup
Ridge (biology) wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Minimal genome wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genome (book) wikipedia , lookup
Oncogenomics wikipedia , lookup
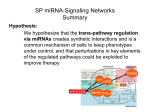
COMPUTATIONAL ANALYSIS OF MULTILEVEL OMICS DATA FOR THE ELUCIDATION OF MOLECULAR MECHANISMS OF CANCER Presented by Azeez Ayomide Fatai Supervisor: Junaid Gamieldien Note: You only have 10-15 minutes maximum, so I suggest presenting only an introduction + section 2 INTRODUCTION Pre-genomic era Post-genomic era Cloning genes at the site of proviral integration Functional assays Positional cloning Tools in clinic MammaPrint Oncotype DX Breast cancer profiling test (HOXB13/IL17RB) Cancer genomics project & Databases TCGA ICGC High-throughput technologies WES and NGS DNA methylation Genomic hybrization Copy number alteration Gene expression profiling DNA methylation Simultaneous study on a cohort of samples Underlying mechanisms Prognostic and predictive biomarkers Target identification Breakdown of my study 1. Network-based identification of candidate cancer genes • Identification of functionally relevant genes in copy number regions • Co-expression and transcriptional analysis 2. Identification of differentially expressed miRNAs and their target genes in the GBM network 3. Identification of prognostic miRNAs for progression-free survival prediction 4. Identification of prognostic protein coding transcripts? genes for progression-free survival prediction 5. Pathway-based and machine learning based feature selection (describe more completely) Identification of differentially expressed miRNAs and their targets in the GBM network INTRODUCTION • Discuss the aims and objectives and the rationale of this section here • State your hypothesis Flowchart for miRNA analysis in GBM Materials and Methods • Add a slide that gives specific details of the method used to identify differentially expressed miRNAs (and WHY they were chosen) • R modules • Underlying statistical tests • p-value cutoffs • fold-change cutoffs (if any) • Describe the samples – numbers, classes, etc • etc Differentially expressed miRNAs between tumour and non-neoplastic brain samples Is there any way to rank these and then list only the ‘best’? Also, be careful to explain what the red text is highlighting Convert the underxpressed fold change as follows: -1/foldchange - that will make 0.1 = -10 fold change for example …continues Produce a better layout if possible – Also highlight any known cancer related miRNAs and genes Any known important genes that you can point out to the audience? Very important: stress that the agreement between miRNA and mRNA expression direction illustrate that the experimental data (and conclusions) are trustworthy Underexpressed miRNA-overexpressed gene network Highlight any known cancer related miRNAs and genes. Also, are there any miRNAs that appear to be regulatory ‘hubs’ based on number of genes they interact with? If so, point them out. Overexpressed miRNA-underexpressed gene network Pathways enriched with miRNA target genes Discussion • What did you learn from this section? • Find anything important? • Eg. is there any disregulated miRNA that looks like it plays dominant major role? • Can it be a drug target? • Is there any gene that can be a drug target? • Etc Conclusions • Biological take home message (e.g. miRNAmRNA networks play a role in GBM… etc) • Mention what you took from this chapter into the next chapters and just give a BRIEF verbal description of the predictive features you found (just to show again that this is just part of a bigger study) Acknowledgements • Your university that sponsors your PhD • Anyone other than me that helped you with data or analysis or tips/clues even in the smallest way • Etc