* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 8 Cell Tour 9 16 05
Survey
Document related concepts
Cellular differentiation wikipedia , lookup
Cell culture wikipedia , lookup
Cell encapsulation wikipedia , lookup
Cell growth wikipedia , lookup
Cytoplasmic streaming wikipedia , lookup
SNARE (protein) wikipedia , lookup
Extracellular matrix wikipedia , lookup
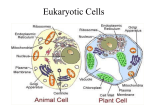
Organ-on-a-chip wikipedia , lookup
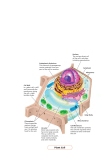
Cell nucleus wikipedia , lookup
Signal transduction wikipedia , lookup
Cytokinesis wikipedia , lookup
Cell membrane wikipedia , lookup
Transcript
A Tour of the Cell Friday Sept 16, 2005 Common features of all cells BCOR 011 Lecture 8 Plasma Membrane – defines inside from outside 1 2 10 µm Plasma membrane Common features of all cells – Functions as a selective barrier – Specific portals for selective transport of Plasma Membrane – defines inside from outside materials in and out of cell Outside of cell Cytosol - Semifluid “inside” of the cell Carbohydrate side chain Hydrophilic region Inside of cell DNA “chromosomes” - Genetic material – hereditary instructions 0.1 µm Hydrophobic region (a) Figure 6.8 A, B TEM of a plasma membrane. The plasma membrane, here in a red blood cell, appears as a pair of dark bands separated by a light band. Hydrophilic region Phospholipid Proteins (b) Structure of the plasma membrane 3 Ribosomes – “factories” to synthesize proteins 4 Two Broad Classes of Cells Cytosol Free ribosomes ER – Carry out protein synthesis Membrane Bound ribosomes Proteins Large To be exported subunit Figure 6.11 Prokaryotes Eukaryotes Pro = before Eu = true karyon = nucleus HAVE A NUCLEUS DO NOT HAVE A NUCLEUS membrane-bound organelles NO internal membranes TEM showing ER and ribosomes 0.5 µm Ribosome – RNA & Protein Complex 5 Small subunit bacteria, cyanobacteria archaebacteria Diagram of a ribosome Plants, Animals, 6 Fungi, protists Pili: attachment structures on the surface of some prokaryotes Nucleoid: region where the cell’s DNA is located (not enclosed by a membrane) Ribosomes: organelles that synthesize proteins Plasma membrane: membrane enclosing the cytoplasm Cell wall: rigid structure outside the plasma membrane No internal membranes Bacterial Cell (Prokaryotic) 7 Bacterial chromosome (a) A typical rod-shaped bacterium Figure 6.6 A, B Capsule: jelly-like outer coating of many prokaryotes 0.5 µm Flagella: locomotion organelles of some bacteria (b) A thin section through the bacterium Bacillus coagulans (TEM) 8 Relative Sizes On the same size scale: “Typical” Bacterium Bacterial cell (Prokaryotic Animal Cell 9 (Eukaryotic) Internal membrane-bound organelles ~ 1-2 µM “Typical” Animal Cell ~ 5 to 20 µM diameter “Typical” Plant Cell ~ 5 to 50 µM diameter µM = micrometer or micron =10-6 meter10 Why Internal Membranes? Compartmentalization (Division of Labor) I’m I’m I’m I’m playing watching cooking sleeping my TV dinner sax Animal Cell (Eukaryotic) 11 12 Animal Cell endoplasmic reticulum ENDOPLASMIC RETICULUM (ER) Rough ER nucleus NUCLEUS Smooth ER Nucleus: Information storage double membrane “Nuclear Envelope” Plasma membrane Centrosome cytosol CYTOSKELETON nucleolus Microfilaments Intermediate filaments Ribosomes ribosomes Microtubules Golgi apparatus Golgi apparatus Peroxisome Figure 6.9 mitochondrion Mitochondrion Lysosome lysosome In animal cells but not plant cells: Lysosomes Centrioles Flagella (in some plant sperm) 13 The NUCLEUS Double membrane Nuclear pores Nuclear Lamina DNA housed, copied, read nuclear envelope Nucleus Nucleus 1 µm Nucleolus 14 Nucleolus Chromatin Nuclear envelope: Inner membrane Outer membrane DNA RNA protein lipid (membrane) Euchromatin Heterochromatin Nuclear pores Pore complex Rough ER Surface of nuclear envelope. 1 µm Ribosome 0.25 µm Close-up of nuclear envelope 15 Figure 6.10 Pore complexes (TEM). Nuclear lamina Nuclear lamina (TEM). 16 Nucleolus Site of Euchromatin region Site of mRNA synthesis Ribosome Subunit Assembly Expression Of Informational RNAs Note: No membrane 17 18 Endoplasmic reticulum (ER) Endoplasmic reticulum (ER) [Reticulum – network] Continuous network of flattened sacs tubules, vesicles, throughout eukaryotic cytoplasm Smooth ER Smooth ER – – – – Rough ER 1 :m 19 Synthesizes membrane lipids Synthesizes steroids Stores calcium Detoxifies poison 20 Example: detoxification in smooth ER Rough ER – Benzo(a)pyrene charred meat, cigarette smoke ribosomes attached to cytoplasmic face • Large flattened sheets • Synthesizes secreted proteins, membrane proteins Oxidations – more soluble exported • Protein modification; initial steps of carbohydrate addition - glycoproteins Some metabolites are more toxic Chronic use of barbiturates, alcoholSER proliferation, resistance 21 22 1 Nuclear envelope is connected to rough ER, which is also continuous with smooth ER Nucleus Rough ER Rough ER 2 Membranes and proteins produced by the ER flow in the form of transport vesicles to the Golgi Slips proteins Through ER membrane Glycosylation Adds oligosaccharides added as protein being made 3 Smooth ER cis Golgi Golgi pinches off transport Vesicles and other vesicles that give rise to lysosomes and Vacuoles Plasma membrane trans Golgi 23 Figure 6.16 4 Lysosome available for fusion with another vesicle for digestion 5 Transport vesicle carries proteins to plasma membrane for secretion 6 Plasma membrane 24 expands by fusion of vesicles; proteins are secreted from cell Golgi Apparatus: protein secretion Processing, packaging and sorting center Cis Golgi Close To RER Functions of the Golgi Apparatus cis Golgi “near” - processing center Present wrapping Service – Trans Golgi modifies proteins trans Golgi - sorting center Far side Away From RER “far” Fed Ex Central Sorts for delivery To specific 26 compartments 25 Functions of the Golgi Apparatus Molecular tags route proteins to proper destination •Trimming of Oligosaccharide side chains on glycosylated proteins •Addition of new Oligosaccharide residues to existing side chains of glycosylated proteins P added in cis Golgi •“Maturation” Cleavages of specific proteins Proteins with M-6-P tag bind receptor in trans Golgi e.g., insulin •Phosphorylation of specific sugar residues on oligosaccharide side chains of glycosylated proteins “molecular zip codes” 27 28 Lysosomes: “Recycling Center” 1 µm Nucleus sacs of digestive enzymes Endocytosis And Phagocytosis Lysosome Hydrolytic enzymes digest food particles Food vacuole fuses with lysosome Lysosome contains active hydrolytic enzymes Digestive enzymes Lysosome Plasma membrane Digestion Food vacuole 29 Figure 6.14 A 30 (a) Phagocytosis: lysosome digesting food In phagocytosis, a cell engulfs a particle by Wrapping pseudopodia around it and packaging it within a membraneenclosed sac large enough to be classified as a vacuole. The particle is digested after the vacuole fuses with a lysosome containing hydrolytic enzymes. PHAGOCYTOSIS EXTRACELLULAR CYTOPLASM FLUID Pseudopodium 1 µm Pseudopodium of amoeba “Food” or other particle Bacterium Food vacuole Food vacuole An amoeba engulfing a bacterium via phagocytosis (TEM). In pinocytosis, the cell “gulps” droplets of extracellular fluid into tiny vesicles. It is not the fluid itself that is needed by the cell, but the molecules dissolved in the droplet. Because any and all included solutes are taken into the cell, pinocytosis is nonspecific in the substances it transports. PINOCYTOSIS 0.5 µm Plasma membrane Pinocytosis vesicles forming (arrows) in a cell lining a small blood vessel (TEM). Receptor-mediated endocytosis enables the cell to acquire bulk quantities of specific substances, even though those substances may not be very concentrated in the extracellular fluid. Embedded in the membrane are proteins with specific receptor sites exposed to the extracellular fluid. The receptor proteins are usually already clustered in regions of the membrane called coated pits, which are lined on their cytoplasmic side by a fuzzy layer of coat proteins. Extracellular substances (ligands) bind to these receptors. When binding occurs, the coated pit forms a vesicle containing the ligand molecules. Notice that there are relatively more bound molecules (purple) inside the vesicle, other molecules (green) are also present. After this ingested material is liberated from the vesicle, the receptors are recycled to the plasma membrane by the same vesicle. RECEPTOR-MEDIATED ENDOCYTOSIS Coat protein Receptor Coated vesicle Ligand Coated pit A coated pit and a coated vesicle formed during receptormediated endocytosis (TEMs). Coat protein Vesicle Plasma membrane Figure 7.20 0.25 µm 31 32 Lysosome containing two damaged organelles 1µm Vesicles move thru the endomembrane system Mitochondrion fragment • Autophagy exocytosis Peroxisome fragment Lysosome fuses with vesicle containing damaged organelle Hydrolytic enzymes digest organelle components Lysosome Digestion Figure 6.14 B Vesicle containing damaged mitochondrion endocytosis 33 34 (b) Autophagy: lysosome breaking down damaged organelle Mitochondria: Powerhouses of the cell Mitochondria singular = mitochondrion •powerhouse of the animal cell produces ~ 90% of ATP •Carries out oxidative reactions •Believed Derived from prokaryotic ancestor 35 - DNA - ribosomes - double membrane – inner and outer *define two functional spaces 36 Mitochondria are enclosed by two membranes – A smooth outer membrane – An inner membrane folded into cristae Gel Important chemical reactions cytoskeleton - eukaryotes Mitochondrion Intermembrane space Outer membrane Free ribosomes in the mitochondrial matrix Cell – organelles = Cytosol Inner membrane Cristae Figure 6.17 Mitochondrial DNA Matrix 37 100 µm 38 Microtubules Microfilaments Intermediate • The cytoskeleton – – – Is a network of fibers extending throughout the cytoplasm Structural Support Movement of Materials and Organelles There are three types of fibers that make up the cytoskeleton Microtubule Tubulin 25 µM dia Cell shape Organelle movt Chromosome separation Flagellar movt Motors: Dynein Kinesis Figure 6.20 0.25 µm Actin 7 µM dia Filaments various 8-15 µM dia Cell shape Cell cleavage Cytoplasmic streaming Muscle contract Nuclear lamina Tension bearing elements Anchors Motors: Myosin Microfilaments 39 40 Table 6.1 – Movement of Vesicles along Microtubules Motor MAPs transport vesicles Vesicle ATP Receptor for motor protein Motor protein (ATP powered) Microtubule of cytoskeleton (a) Motor proteins that attach to receptors on organelles can “walk” the organelles along microtubules or, in some cases, microfilaments. Vesicles Microtubule Figure 6.21 A, B (b) Vesicles containing neurotransmitters migrate to the tips of nerve cell axons via the mechanism in (a). In this SEM of a squid giant axon, two vesicles can be seen moving along a microtubule. (A separate part of the experiment provided the evidence that they were in fact moving.) Dynein inbound 0.25 µm outbound kinesin MTOC 41 42 • Animal cells – Lack cell walls – Are covered by an elaborate matrix, the ECM – Contains a pair of centrioles • The ECM Is made up of glycoproteins Centrosome EXTRACELLULAR FLUID Collagen A proteoglycan complex Microtubule Polysaccharide molecule Carbohydrates Centrioles Core protein 0.25 µm Fibronectin “microtubuleorganizing center” Figure 6.22 Longitudinal section of one centriole Microtubules Cross section of the other centriole Plasma membrane Integrin 43 Figure 6.29 Integrins Microfilaments Proteoglycan molecule CYTOPLASM 44 plant cell Ribosomes (small brown dots) Rough endoplasmic reticulum • Functions of the ECM include – Cell-Cell adhesion – Cell-Cell recognition – Regulation of cellular processes Smooth endoplasmic reticulum NUCLEUS Golgi apparatus Central vacuole/Tonoplast Microfilaments Intermediate filaments CYTOSKELETON Microtubules Mitochondrion Peroxisome Plasma membrane Chloroplast Cell wall Wall of adjacent cell 45 Plant Central vacuoles - Tonoplasts – – – Plasmodesmata Figure 6.9 46 In plant cells, chloroplasts capture energy from the sun Are found in plant cells Hold reserves of important organic compounds and water Regulates Turgor Chloroplast Central vacuole Photosynthesis Ribosomes Stroma Cytosol Chloroplast DNA Inner and outer membranes Tonoplast Granum Nucleus Central vacuole 1 µm Cell wall Thylakoid Chloroplast Figure 6.15 Figure 6.18 47 5 µm 48 3 Central Players Chloroplasts Inner Chlorplast Membrane OuterChlorplast Membrane Stroma -Contain DNA -Contain bacterial-like ribosomes -Believed derived from prokaryotic ancestor Thylakoid Space cyanobacterium = blue-green alga -Double membrane organelle defines three functional spaces 49 Thylakoid Membrane Intermembrane Space (transports things in and out of the chloroplast, but not central to photosynthesis itself 50 Cell Walls of Plants • The cell wall – Is an extracellular structure of plant cells that distinguishes them from animal cells 51 52 • Plant cell walls – Are made of cellulose fibers embedded in other polysaccharides and protein – May have multiple layers Plasma membrane Central vacuole of cell Secondary cell wall • Plasmodesmata – Are channels that perforate plant cell walls Cell walls Primary cell wall Central vacuole of cell Middle lamella Interior of cell 1 µm Central vacuole Cytosol Plasma membrane Plant cell walls Interior of cell Figure 6.30 Figure 6.28 Plasmodesmata 53 Summary Features of all cells Features of Prokaryotes Organelles of Animal Cells Organelles of Plant Cells 55 0.5 µm Plasmodesmata Plasma membranes 54