* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download AP_Gene to Protein
Gene desert wikipedia , lookup
Protein moonlighting wikipedia , lookup
Genome evolution wikipedia , lookup
Community fingerprinting wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Bottromycin wikipedia , lookup
Biochemistry wikipedia , lookup
Molecular evolution wikipedia , lookup
List of types of proteins wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Polyadenylation wikipedia , lookup
Point mutation wikipedia , lookup
RNA silencing wikipedia , lookup
RNA interference wikipedia , lookup
Gene regulatory network wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Expanded genetic code wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Non-coding RNA wikipedia , lookup
Genetic code wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Transfer RNA wikipedia , lookup
Messenger RNA wikipedia , lookup
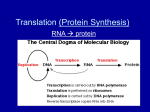
I. Genes & Proteins ● Archibald Garrod (1909): first to contend that genes specified enzymes that, in turn, controlled phenotypic traits. This hypothesis was experimentally verified in the 1940’s by the work of Beadle & Tatum (see fig. 1). Figure 1: Beadle & Tatum’s Experiment To test the one-gene, one enzyme hypothesis, Beadle & Tatum exposed the fungus Neruospora to radiation in order to mutate genes involved in the synthesis of the essential amino acid arginine. If this hypothesis was correct, each mutation would affect a SINGLE gene in the arginine pathway, resulting in the lack of a SINGLE enzyme coded for by that gene: *Minimal Medium (MM): provides all of the essential raw materials to produce arginine (assuming the pathway is not disrupted via mutation) To identify specific defect in each class of mutant, Beadle & Tatum distributed them to a number of vials, each with minimal medium + a single nutrient. The particular supplement that allowed growth indicated the metabolic defect. These results showed that each mutant had a mutation in a SINGLE gene & that each gene affected only ONE enzyme. ● ● Beadle & Tatum (One-Gene, One Enzyme Hypothesis): a) Researchers later realized that genes not only code for enzymes, but other proteins as well. Thus they began to think in terms of one-gene, one protein. However, many proteins are constructed from 2 or more different polypeptides, each specified by its own gene. Therefore, Beadle & Tatum’s idea can be restated as the One-Gene, One-Polypeptide Hypothesis. 1 II. Gene Expression & RNA Function Figure 2: Central Dogma The Central Dogma of molecular biology was originally proposed by Francis Crick in 58’ to suggest the possible relay of genetic information in the cell. Was later experimentally verified. ● ● Gene Expression: Advantages of RNA Intermediates 1) The DNA (& genetic code) can remain protected from destructive enzymes & free radicals within the nucleus. 2) The RNA intermediate can be “edited” in a variety of ways to form multiple, related protein products without altering the genetic code within the DNA itself: mRNA editing reduces the number of genes to produce all of the proteins required to maintain cellular metabolism. 3) In prokaryotes, multiple ribosomes can interact with a single mRNA molecule to maximize the amount of protein formed from a gene: 2 RNA Structure Figure 3: RNA vs DNA RNA nucleotides contain the 5-sided sugar Ribose (-OH on 2’ carbon of sugar) instead of deoxyribose. May bear the bases adenine, guanine, cytosine, & Uracil on the sugar’s 1’ carbon. ● ● Types a) b) c) d) of RNA differ in abundance, structure, function & size: rRNA: most abundant (80%) & largest of all varieties of RNA (120-3500bp). tRNA: comprise 15% of all cellular RNA; size ranges between 75-90bp. mRNA: least abundant (5%); size is variable. miRNA & siRNA: function in regulating gene expression via RNAi (i = “interference”). II. Eukaryotic Gene Structure Figure 4: Organization of Eukaryotic Genes 3 The Promoter, located upstream from the start end of every eukaryotic gene, contains the base sequence TATAAT or TATA Box that functions to signal the start end of the gene. Consequently, the enzyme & proteins responsible for the transcription process can “find” the gene & bind to this region. ● The region of a gene containing the stretch of DNA that codes for a polypeptide is called a Transcriptional Unit. At the end of the transcriptional unit is a Terminator sequence that signals the end of the gene sequence. ● Figure 4.1: Organization of Eukaryotic Genes: Codons ● Exons a) The nucleotide bases within an exon are organized into triplets called a Codon. Each codon dictates a single amino acid of the polypeptide product of the gene. ● Introns a) Introns allow for mRNA to be edited in a variety of different ways, leading to the formation of multiple gene products from the SAME gene sequence. III. Gene Expression: Transcription ●The first step in the expression of the information encoded by a gene into a protein product is Transcription. In this process, the base sequence of a gene is used to create a complementary molecule of mRNA. Once formed, the mRNA can leave the nucleus (“mobile gene”) & interact with a ribosome in the cytosol, whereupon the instructions it carries will be translated to form a polypeptide protein. ● 4 Figure 5: Transcription: Initiation Phase ● Initiation a) Some of the transcription factors that associate with RNA polymerase II at the gene promoter act like helicase to separate the strands of the gene. b) The strand running from the 3’ to the 5’ direction is the Sense Strand, acting as the template off of which complimentary mRNA is polymerized. Figure 5.1: Transcription: Elongation Phase 5 ● Elongation a) During this process, RNA polymerase II adds nucleotides complimentary to the DNA template (sense) strand, elongating the mRNA in its 3’ direction. b) As the mRNA elongates, its 5’ end falls away from the template, allowing the separated DNA strands behind it to rewind into a helix (thus only one section of the gene is ever exposed at any one time). Figure 5.2: Transcription: Termination Phase The 5’ UTR has recognition signals for ribosome binding to properly position ribosomes to translate the genetic code. The Reading Frame, located immediately downstream from the leader, contains the actual message for the polypeptide. Within this region, codons specify a single amino acid for the polypeptide to be made. ● A Start Codon (AUG) signals the beginning of the coding sequence whereas a Stop Codon (UAA, UAG, UGA) signals the end. Stop codons are followed by a noncoding 3’ UTR. ● ● Termination a) During prokaryotic termination, RNA polymerase IMMEDIATELY detaches from the template upon reaching the terminator sequence. In eukaryotes, transcription continues for another 20-30 bp’s after reaching the terminator. 6 Figure 6: mRNA Reading Frame: Codons Messenger RNA Processing Figure 7: RNA Processing: Addition of 5’ Cap When the primary mRNA transcript is 20-30 nucleotides long, its 5’ end is capped with a form of the guanine nucleotide. This 5’ CAP protects the mRNA from degradation by hydrolytic exonucleases. Furthermore, it serves as a binding site for ribosomes. ● 7 Figure 7.1: RNA Processing: Addition of Poly-A Tail To the 3’end is attached a Poly-A Tail consisting of 30-200 adenine nucleotides. The poly-A tail inhibits degradation by hydrolytic exonucleases & aids in ribosome attachment. It also seems to aid in the export of mRNA from the nucleus. ● Figure 7.2: mRNA Processing: Intron Removal RNA polymerase transcribes both introns & exons. Particles called Small Nuclear Ribonuclearprotiens (“snurps”) join with additional proteins to form a Spliceosome. This complex locates & excises (cuts) introns from the primary ● 8 transcript & splices the exons together to form a final transcript that is much shorter than the original gene sequence (& more easily handled by the ribosome). Figure 7.3: Differential mRNA Splicing ● Alternative mRNA Splicing: a) Alternative mRNA splicing leads to the production of multiple proteins from a single gene & may be one reason humans can get along with a relatively small number of genes (20-25,000). III. Gene Expression: Translation ●The final step in the expression of the information encoded by a gene into a protein product is Translation. In this process, the base sequence of mRNA is deciphered w/in a ribosome to create a polypeptide/protein with assistance from tRNA. Figure 8: Transfer RNA (tRNA) 9 ● tRNA: a) The amino acid binding site is at the 3’ end of the molecule, called the Amino Acid Binding Site; the carboxyl group of the amino acid is bound to the exposed OH group of the tRNA, leaving the amino group of the amino acid free to participate in peptide bond formation. b) Amino acids are added to the 3’ end of tRNA by an Aminoacyl-tRNA Synthase, after which the tRNA is regarded as being “charged” (can actively participate in protein synthesis). Figure 8.1: Transfer RNA: Anticodon Three loops of unpaired nucleotides exist w/in tRNA, one of which contains a sequence called an Anticodon, which is complementary to certain codons on mRNA. The anticodon enables the genetic code to be deciphered, whereby appropriate amino acids are transferred to the ribosome in a specific sequence to synthesize a polypeptide. ● Figure 8.2: Transfer RNA Anticodon: Wobble Base Position The base occupying the 5’ position of an anticodon may not obey conventional base-paring rules. This “wobble” base has the tendency to pair with multiple bases at the 3’ position of an mRNA codon… ● 10 Figure 8.3: Transfer RNA Anticodon: Wobble Base Pairing & “Silent” Mutations *Since the mutation at the 3’ position of the mRNA codon (GGC GGU) corresponds to the wobble position of the anticodon, its amino acid “meaning” will not change. ● Wobble Position: Figure 9: Ribosome Structure & Function: Overview In prokaryotes & eukaryotes, the large & small subunits join to form a functional protein only when they attach to mRNA. During translation, the mRNA fits in a groove between the contact surfaces of the two subunits. ● ● In addition to the binding site for mRNA, ribosomes have 3 binding sites for tRNAs: a) P Site: holds the tRNA carrying the growing polypeptide chain. b) A Site: holds the tRNA arrying the next amino acid in the chain. c) E Site: site from which discharged tRNAs leave the ribosome. 11 ● Ribosomes: Figure 10: Translation: Initiation Phase (Early) 5’ CAP of mRNA serves as a “landing platform” for the attachment of the small (30s) ribosome subunit. 30s subunit “scans” mRNA for the start codon, whereupon the initiator tRNAmet binds to it. ● Figure 101: Translation: Initiation Phase (Late) The consumption of GTP promotes the addition of the large (50s) subunit, whereupon the initiator tRNAmet resides in the ribosome’s P site. Association of tRNA, mRNA, & 30/50s subunits is dependent on a host of Translation Initiation Factors. ● 12 Figure 10.2: Elongation & Translocation During Elongation, amino acids are added one by one to the initial amino acid (methionine). An anticodon on tRNA “reads” the codon of mRNA exposed in the ribosome’s A site. If complementary, the amino acid carried by the tRNA will form a peptide bond with the amino acid of the tRNA occupying the P site. ● During Translocation, a sub-stage of elongation, the tRNA in the P site dissociates from the ribosome, & the tRNA in the A site, carrying the growing polypeptide, is shifted to the P site. This movement brings into the A site the next codon to be translated on the mRNA. ● Figure 10.3: Translation: Termination Phase Elongation continues until a stop codon (UAA, UAG, & UGA) reaches the A site of the ribosome. A protein called a Release Factor binds to the termination codon in the A site, causing the release of the polypeptide chain from the tRNA. This hydrolysis initiates the dissociation of the ribosomal subunits as well as the release of the mRNA transcript. ● 13 The liberated polypeptide chains will undergo a modification process as amino acids may be altered by the addition of sugars, lipids, phosphate groups, etc. The chain will assume a specific conformation (3-D shape) with the help of molecular chaperones & join with other polypeptide chains to form a functional protein or enzyme. ● IV. Regulating Gene Expression: RNA Interference (RNAi) Figure 11: RNAi: General Mechanism RNAi via Micro-RNA (miRNA) or Small Interfering RNA (siRNA) acts to “capture” mRNA in route to a ribosome prior to being translated. Consequently, no protein product is made & gene expression is essentially blocked. Functions of RNAi within the cell includes: a) RNAi participates in silencing of certain viral mRNA’s to block their translation into viral proteins. Thus, RNAi can inhibit the reproduction of certain viral strains to stop further infection. b) RNAi participates in Gene Silencing: mi/siRNA genes are selectively active in some cell types, leading to differential cell development. c) miRNA / RISC complex can intercept mRNA from harmful genes, even those of cancer cells to block their spread (e.g. angiogenesis gene, telomerase gene, etc) ● 14