* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Bioinformatics Tools
Polycomb Group Proteins and Cancer wikipedia , lookup
Protein moonlighting wikipedia , lookup
Copy-number variation wikipedia , lookup
Mitochondrial DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Point mutation wikipedia , lookup
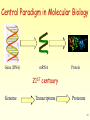
Vectors in gene therapy wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
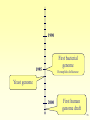
Oncogenomics wikipedia , lookup
Transposable element wikipedia , lookup
Genetic engineering wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Gene expression profiling wikipedia , lookup
Microevolution wikipedia , lookup
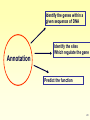
Genome (book) wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Designer baby wikipedia , lookup
Metagenomics wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Non-coding DNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genomic library wikipedia , lookup
History of genetic engineering wikipedia , lookup
Human genome wikipedia , lookup
Pathogenomics wikipedia , lookup
Minimal genome wikipedia , lookup
Helitron (biology) wikipedia , lookup
Public health genomics wikipedia , lookup
Human Genome Project wikipedia , lookup
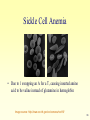
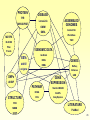
Introduction to Bioinformatics 234525-236523 Lecturer: Dr. Yael Mandel-Gutfreund Teaching Assistance: Martin Akerman Sivan Bercovici Course web site : http://webcourse.cs.technion.ac.il/234525 What is Bioinformatics? 2 Course Objectives • To introduce the bioinfomatics discipline • To make the students familiar with the major biological questions which can be addressed by bioinformatics tools • To introduce the major tools used for sequence and structure analysis and explain in general how they work (limitation etc..) 3 Course Structure and Requirements 1.Class Structure 1. 2. 2 hours Lecture 1 hour tutorial 2. Home work • Homework projects will be given every second week • The homework will be done in pairs. • 5/5 homework projects submitted 2. A final project will be conducted and submitted in pairs 4 Grading • 30 % Homework assignments • 70% final project 5 Literature list • Gibas, C., Jambeck, P. Developing Bioinformatics Computer Skills. O'Reilly, 2001. • Lesk, A. M. Introduction to Bioinformatics. Oxford University Press, 2002. • Mount, D.W. Bioinformatics: Sequence and Genome Analysis. 2nd ed.,Cold Spring Harbor Laboratory Press, 2004. Advanced Reading Jones N.C & Pevzner P.A. An introduction to Bioinformatics algorithms MIT Press, 2004 6 What is Bioinformatics? 7 What is Bioinformatics? “The field of science in which biology, computer science, and information technology merge to form a single discipline” Ultimate goal: to enable the discovery of new biological insights as well as to create a global perspective from which unifying principles in biology can be discerned. 8 from purely lab-based science to an information science Bioinformatics Bio = Informatics 9 Central Paradigm in Molecular Biology Gene (DNA) mRNA Protein 21ST centaury Genome Transcriptome Proteome 10 Genome • Chromosomal DNA of an organism • Coding and non-coding DNA • Genome size and number of genes does not necessarily determine organism complexity 11 Transcriptome • Complete collection of all possible mRNAs (including splice variants) of an organism. • Regions of an organism’s genome that get transcribed into messenger RNA. • Transcriptome can be extended to include all transcribed elements, including non-coding RNAs used for structural and regulatory purposes. 12 Proteome • The complete collection of proteins that can be produced by an organism. • Can be studied either as static (sum of all proteins possible) or dynamic (all proteins found at a specific time point) entity 13 From DNA to Genome Watson and Crick DNA model First protein sequence 1955 1960 First protein structure 1965 1970 1975 1980 1985 14 1990 First bacterial genome 1995 Hemophilus Influenzae Yeast genome 2000 First human genome draft 15 Complete Genomes Total 2008 2007 706 456 Eukaryotes 78 43 Bacteria 578 383 Archaea 50 29 16 Perhaps not surprising!!! How humans are chimps? Comparison between the full drafts of the human and chimp genomes revealed that they differ only by 1.23% 17 What’s Next ? The “post-genomics” era Annotation Comparative genomics Structural genomics Functional genomics Goal: to understand the living cell 18 Annotation CCTGACAAATTCGACGTGCGGCATTGCATGCAGACGTGCATG CGTGCAAATAATCAATGTGGACTTTTCTGCGATTATGGAAGAA CTTTGTTACGCGTTTTTGTCATGGCTTTGGTCCCGCTTTGTTC AGAATGCTTTTAATAAGCGGGGTTACCGGTTTGGTTAGCGAGA AGAGCCAGTAAAAGACGCAGTGACGGAGATGTCTGATG CAA TAT GGA CAA TTG GTT TCT TCT CTG AAT ...... .............. TGAAAAACGTA 19 Identify the genes within a given sequence of DNA Identify the sites Which regulate the gene Annotation Predict the function 20 TF binding site CCTGACAAATTCGACGTGCGGCATTGCATGCAGACGTGCATG CGTGCAAATAATCAATGTGGACTTTTCTGCGATTATGGAAGAA CTTTGTTACGCGTTTTTGTCATGGCTTTGGTCCCGCTTTGTTC AGAATGCTTTTAATAAGCGGGGTTACCGGTTTGGTTAGCGAGA AGAGCCAGTAAAAGACGCAGTGACGGAGATGTCTGATG CAA TAT GGA CAA TTG GTT TCT TCT CTG AAT ................................. Transcription Start Site promoter .............. TGAAAAACGTA ORF=Open Reading Frame Ribosome binding Site CDS=Coding Sequence 21 Comparative genomics Human ATAGCGGGGGGATGCGGGCCCTATACCC Chimp ATAGGGG - - GGATGCGGGCCCTATACCC Mouse ATAGCG - - - GGATGCGGCGC -TATACCA 22 Researchers have learned a great deal about the function of human genes by examining their counterparts in simpler model organisms such as the mouse. Conservation of the IGFALS (Insulin-like growth factor) Between human and mouse. 23 Functional genomics 24 Understanding the function of genes and other parts of the genome 25 A network of interactions can be built For all proteins in an organism A large network of 8184 interactions among 4140 S. Cerevisiae proteins 26 Structural genomics 27 Assigning the structures of all proteins protein complexes Biologic processes Shape and electrostatics Active sites fold Evolutionary relationship Protein-ligand complexes Functional sites 28 Resources and Databases The different types of data are collected in database – Sequence databases – Structural databases – Databases of Experimental Results All databases are connected 29 Sequence databases • • • • Gene database Genome database SNPs database Disease related mutation database 30 Gene database • Give information into gene functionality • Alternative splicing of genes – Alternative pattern of exons included to create gene product • EST 31 Genome Databases • Data organized by species • Clones assembled into contigous pieces ‘contigs’ or whole chromosomes • Information on non-coding regions • Relativity 32 Genome Browsers • Annotation adds value to sequence • Easy “walk” through the genome • Comparative genomics 33 Genome Browsers • UCSC Genome Browser http://genome.ucsc.edu/ • Ensembl Genome Browser (http://www.ensembl.org) • WormBase: http://www.wormbase.org/ • AceDB: http://www.acedb.org/ • Comprehensive Microbial Resource: http://www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl • FlyBase: http://flybase.bio.indiana.edu/ 34 SNP database Single Nucleotide Polymorphisms (SNPs) • Single base difference in a single position among two different individuals of the same species • Play an important role in differentiation and disease 35 Sickle Cell Anemia • Due to 1 swapping an A for a T, causing inserted amino acid to be valine instead of glutamine in hemoglobin Image source: http://www.cc.nih.gov/ccc/ccnews/nov99/ 36 Healthy Individual >gi|28302128|ref|NM_000518.4| Homo sapiens hemoglobin, beta (HBB), mRNA ACATTTGCTTCTGACACAACTGTGTTCACTAGCAACCTCAAACAGACACCATGGTGCATCTGACTCCTGA GGAGAAGTCTGCCGTTACTGCCCTGTGGGGCAAGGTGAACGTGGATGAAGTTGGTGGTGAGGCCCTGGGC AGGCTGCTGGTGGTCTACCCTTGGACCCAGAGGTTCTTTGAGTCCTTTGGGGATCTGTCCACTCCTGATG CTGTTATGGGCAACCCTAAGGTGAAGGCTCATGGCAAGAAAGTGCTCGGTGCCTTTAGTGATGGCCTGGC TCACCTGGACAACCTCAAGGGCACCTTTGCCACACTGAGTGAGCTGCACTGTGACAAGCTGCACGTGGAT CCTGAGAACTTCAGGCTCCTGGGCAACGTGCTGGTCTGTGTGCTGGCCCATCACTTTGGCAAAGAATTCA CCCCACCAGTGCAGGCTGCCTATCAGAAAGTGGTGGCTGGTGTGGCTAATGCCCTGGCCCACAAGTATCA CTAAGCTCGCTTTCTTGCTGTCCAATTTCTATTAAAGGTTCCTTTGTTCCCTAAGTCCAACTACTAAACT GGGGGATATTATGAAGGGCCTTGAGCATCTGGATTCTGCCTAATAAAAAACATTTATTTTCATTGC >gi|4504349|ref|NP_000509.1| beta globin [Homo sapiens] EEKSAVTALWGKVNVDEVGGEALGRLLVVYPWTQRFFESFGDLSTPDAVMGNPKVKAHGKKVLG MVHLTP AFSDGLAHLDNLKGTFATLSELHCDKLHVDPENFRLLGNVLVCVLAHHFGKEFTPPVQAAYQKVVAGVAN ALAHKYH 37 Diseased Individual >gi|28302128|ref|NM_000518.4| Homo sapiens hemoglobin, beta (HBB), mRNA ACATTTGCTTCTGACACAACTGTGTTCACTAGCAACCTCAAACAGACACCATGGTGCATCTGACTCCTGA GGTGAAGTCTGCCGTTACTGCCCTGTGGGGCAAGGTGAACGTGGATGAAGTTGGTGGTGAGGCCCTGGGC AGGCTGCTGGTGGTCTACCCTTGGACCCAGAGGTTCTTTGAGTCCTTTGGGGATCTGTCCACTCCTGATG CTGTTATGGGCAACCCTAAGGTGAAGGCTCATGGCAAGAAAGTGCTCGGTGCCTTTAGTGATGGCCTGGC TCACCTGGACAACCTCAAGGGCACCTTTGCCACACTGAGTGAGCTGCACTGTGACAAGCTGCACGTGGAT CCTGAGAACTTCAGGCTCCTGGGCAACGTGCTGGTCTGTGTGCTGGCCCATCACTTTGGCAAAGAATTCA CCCCACCAGTGCAGGCTGCCTATCAGAAAGTGGTGGCTGGTGTGGCTAATGCCCTGGCCCACAAGTATCA CTAAGCTCGCTTTCTTGCTGTCCAATTTCTATTAAAGGTTCCTTTGTTCCCTAAGTCCAACTACTAAACT GGGGGATATTATGAAGGGCCTTGAGCATCTGGATTCTGCCTAATAAAAAACATTTATTTTCATTGC >gi|4504349|ref|NP_000509.1| beta globin [Homo sapiens] VEKSAVTALWGKVNVDEVGGEALGRLLVVYPWTQRFFESFGDLSTPDAVMGNPKVKAHGKKVLG MVHLTP AFSDGLAHLDNLKGTFATLSELHCDKLHVDPENFRLLGNVLVCVLAHHFGKEFTPPVQAAYQKVVAGVAN ALAHKYH 38 Disease Databases • Genes are involved in disease • Many diseases are well studied • Description of diseases and what is known about them is stored 39 Structure Databases • 3-dimensional structures of proteins, nucleic acids, molecular complexes etc • 3-d data is available due to techniques such as NMR and X-Ray crystallography 40 41 Databases of Experimental Results • Data such as experimental microarray images- expression data • Proteomic data • Metabolic pathways, protein-protein interaction data, regulatory networks • ETC…………. 42 Literature Databases PubMed http://www.ncbi.nlm.nih.giv/PubMed Service of the National Library of Medicine • MEDLINE publication database – Over 17,000 journals – 15 million citations since 1950 43 Putting it all Together • Each Database contains specific information • Like other biological systems also these databases are interrelated 44 PROTEIN PIR DISEASE ASSEMBLED GENOMES LocusLink SWISS-PROT OMIM GoldenPath OMIA WormBase MOTIFS TIGR BLOCKS Pfam GENOMIC DATA Prosite GenBank ESTs dbEST DDBJ GENES EMBL RefSeq unigene AllGenes SNPs GENE EXPRESSION dbSNP STRUCTURE PDB MMDB SCOP PATHWAY Stanford MGDB KEGG NetAffx COG ArrayExpress GDB LITERATURE PubMed 45