* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Structure and Functions of Ribosomes
Cellular differentiation wikipedia , lookup
Cell membrane wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
Cell culture wikipedia , lookup
Signal transduction wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Extracellular matrix wikipedia , lookup
Cell growth wikipedia , lookup
Cell nucleus wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Endomembrane system wikipedia , lookup
Cytokinesis wikipedia , lookup
Proteolysis wikipedia , lookup
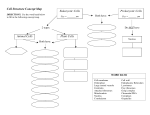
Structure and Functions of Ribosomes Project Assignment S-114.500 Principles for Biosystems of the Cell Mailiina Turanlahti 01/12/2004 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 1 Agenda Introduction Structure of Ribosomes Functions of Ribosomes Research Methods for the study of Ribosomes Programs for Visualisation and Modelling 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 2 Introduction: What are Ribosomes Small cytoplasmic organelles found in all prokaryotes and eukaryotes in large numbers Typically 10,000 ribosomes in a bacterial cell and many more in eukaryotic cells Size: ~20 nm diameter Major actors in protein synthesis Different “versions” 80S* in Eukaryotes 70S in Prokaryotes 55S in eukaryotic chloroplasts and mitochondria May exist in two forms in Eukaryotes Bound to endoplasmic reticulum, forming rough ER Free floating in the cytoplasm * S = Svedberg units: a measure of the rate of sedimentation of a component in a centrifuge that is related both to the molecular weight and the three-dimensional shape of the component 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 3 Agenda Introduction Structure of Ribosomes Functions of Ribosomes Research Methods for the study of Ribosomes Programs for Visualisation and Modelling 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 4 General Features of the Structure of Ribosomes Constitute of a smaller and a larger subunit Building blocks: ribosomal RNA (rRNA) and many proteins Most of the volume is occupied by RNA 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 5 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 6 Prokaryotic Ribosomes Small ribosomal subunits have a head and a base with an armlike platform extending from one side. The large subunit has a prominent central protuberance, stalk, and ridge extending from one side. The large subunit has a tunnel about 10nm long and 2.5 nm in diameter, that extends from the region containing the A (aminoacyl) and P (peptidyl) sites to the part of the large subunit from which the newly assembled polypeptide chain exits the ribosome 01/12/04 Eukaryotic Ribosomes Contains some additional features on the small subunit, including a bill that extends from the head on the side opposite to the cleft a set of lobes at the end of the subunit opposite to the head Has three rRNAs (28S, 5.8S and 5S) on the large subunit, as opposed to two on the prokaryotic one Proteins differ slightly from one organism to another. For instance, that mammals have the largest ribosomes, due to different proteins 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 7 Agenda Introduction Structure of Ribosomes Functions of Ribosomes Research Methods for the study of Ribosomes Programs for Visualisation and Modelling 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 8 The Function of Ribosomes: Protein Synthesis, or Translation Principle: with the ribosome acting as a controlling site, the ordered arrangement of tRNAs along the mRNA strand carrying the genetic message induces the correct order for the corresponding amino acids to form the polypeptide The translation process consists of three sub processes: 1. Formation of the initiation complex 2. Elongation of the polypeptide chain (one repetition of the steps a, b and c for every amino acid incorporated into the protein being synthesized) a) binding of the aminoacyl-tRNA b) peptide bond formation c) translocation 3. Termination 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 9 Translation (1): Initiation 1. 2. 3. 4. 5. A ribosome separates into large and small subunits Met-tRNA combines with GTP in a side reaction involving an initiation factor Met-tRNA is added to the small ribosomal subunit The small subunit is added to the mRNA in a reaction driven by ATP hydrolysis; attachment takes place at the 5' cap of the mRNA; once attached, the small subunits moves or "scans" along the mRNA until it reaches the AUG initiator codon The large ribosomal subunit is added driven by the hydrolysis of GTP brought to the complex with the initiator tRNA; elongation follows 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 10 Translation (2): Elongation 1. Aminoacyl-tRNA binds to the A site 2. Peptide bond formation 3. Peptidyl-tRNA formed at the A site by step two is transferred from the A site to the P site Peptide bond formation: catalyzed on the ribosome by peptidyl transferase a) Adjacent aminoacyl-tRNAs bound to the mRNA at the ribosome b) following peptide bond formation, an uncharged tRNA is in the P site and a dipeptidyl-tRNA in the A site. 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 11 Translation (3): Termination 1. 2. 3. 4. A stop codon is encountered at the A site which causes the release factor to bind to the A site along with GTP instead of aminoacyltRNA The release factor binds to the stop codon and the bond holding the polypeptide chain to the tRNA site at the P site is hydrolyzed, catalyzed by the peptidyl tranferase site of the large subunit. Since there is no amino acid located at the A site, the hydrolysis allows the polypeptide chain to be freed from the ribosome; with the release of the polypeptide, the release factor is ejected from the A site, and the empty tRNA is ejected from the P site Ribosomal components separate 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 12 Agenda Introduction Structure of Ribosomes Functions of Ribosomes Research Methods for the study of Ribosomes Programs for Visualisation and Modelling 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 13 Cryo-EM maps Technique for getting 3D structures of biological molecules from an electron microscope Gathers many images of the same particle in various orientations, and then uses e.g. computed tomography to reconstruct the 3D structure Usually 15 Ångström resolution, but with enough images (hundreds of thousands) resolutions of 7 or 8 Å are attainable Cryo-EM reconstruction of E. coli ribosome at 11.5 Å resolution. (a) Small subunit; (b) large subunit; (c) assembled ribosome Gabashvili, I.S., Penczek, P et al., Solution structure of the E. coli ribosome at 11.5 Å resolution. Cell, Vol. 100, 2000 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 14 NMR Spectroscopy Enables the research of biological macromolecules, e.g. ribosomal proteins The protein must first be cloned, expressed and purified Samples of the protein are enriched with a radioactive isotope of carbon or nitrogen in a predetermined way. A spectrometer is used to record resonations of amines and single atoms that follow the enriched parts of the molecule. The calculation of the protein structure is an optimisation problem, where the minimum energy is what is looked for. The goal is to identify the full range of structures that are consistent with the distance and angle constraints derived from the NMR data, while making sure that the structure has reasonable molecular geometry, and attains the minimum value of the energy function. The method enables researching both the structure and the dynamics of ribosomal proteins There are significant limitations for the size of the molecule to be researched, as the NMR data is highly complex S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 15 01/12/04 X-Ray Crystallography Enables solving structures to atomic resolution The ribosome, constituting of more than 100,000 atoms, is the most complex biological structure that is solved at this resolution Many approximations and guesses have to be made, though Requires first the crystallization of the protein Crystals are built of well ordered elements, which diffract light or hard X-rays in this case, in a characteristic way. The characteristic diffraction pattern allows in principle to determine the precise location of each individual atom within the crystal, e.g. within the ribosomal particle. The most useful methods of research combine X-ray crystallography and Cryo-EM, by “moulding” the X-ray data into the Cryo-EM maps 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 16 Agenda Introduction Structure of Ribosomes Functions of Ribosomes Research Methods for the study of Ribosomes Programs for Visualisation and Modelling S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 17 01/12/04 Programs for Visualisation and Modelling Aim: to find suitable programs to illustrate the 3D-structure of ribosomes and their parts Requirement: support for some file format in which information about ribosome structure was stored, and made available on the Internet The programs were searched on Structural Biology Software Database. Several different structures were viewed with each of the programs Three programs were tested Biodesigner (not very successfully) MDL Chime Cn3D 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 18 MDL Chime Runs in the browser (IE or Netscape 5.0 or higher) as a plug-in Enables watching molecule(s) provided as a PDB file by the author of the viewed web page Prokaryotic ribosome viewed with Chime. The RNA backbone is shown as pink wire, and proteins as beige spheres. 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 19 Cn3D Helper application that allows viewing 3-D structures from NCBI's Entrez retrieval service A search in Entrez retrieval service gave 282 representations of ribosomal structures (most of a single protein, though) ”Simultaneously displays structure, sequence, and alignment ” Structure of the ribosomal 40S subunit from yeast viewed with Cn3D 01/12/04 S114.500-Principles for biocyctems of the cell Mailiina Turanlahti 20