* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download genetic variation in isoniazid metabolism genes
Genetic drift wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Oncogenomics wikipedia , lookup
Human genome wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Behavioural genetics wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Gene expression programming wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Genetic testing wikipedia , lookup
Gene desert wikipedia , lookup
Non-coding DNA wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Helitron (biology) wikipedia , lookup
Population genetics wikipedia , lookup
Genome editing wikipedia , lookup
Gene therapy wikipedia , lookup
Genome evolution wikipedia , lookup
Medical genetics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genetic engineering wikipedia , lookup
Genome-wide association study wikipedia , lookup
Heritability of IQ wikipedia , lookup
History of genetic engineering wikipedia , lookup
Human genetic variation wikipedia , lookup
Designer baby wikipedia , lookup
Genome (book) wikipedia , lookup
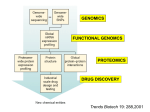
Pharmacogenomics wikipedia , lookup
PHARMACOGENETIC ANALYSIS OF AN ISONIAZID METABOLISM GENE, N-ACETYLTRANSFERASE 2 So Yamada1, Mila Tang2, Suzanne Moadebi2, Julius Halaschek-Wiener1, J. Mark Fitzgerald2, Kevin Elwood2, Fawziah Marra2* and Angela Brooks-Wilson1* 1 Genome Sciences Centre, British Columbia Cancer Agency and 2BC Centre for Disease Control; Vancouver, BC, Canada; *These authors co-led the study BACKGROUND: Pulmonary tuberculosis (TB) is among the most serious public health problems in both developing and developed countries. Incidence rates are increasing in high-risk populations within Canada. The current treatment of latent TB generally includes the administration of isoniazid (INH), a drug known to cause hepatotoxicity as a potentially serious side effect. INH-induced hepatotoxicity derives from toxic metabolites produced during INH breakdown. Genetic polymorphisms in Nacetyltransferase 2 (NAT2), a core enzyme in INH metabolism, have been previously established to play a significant role in the development of hepatotoxicity. The phenotypic response to INH is partly but not entirely determined by the NAT2 genotype. This investigation of the pharmacogenetic effect of NAT2 variants is part of a larger study that will also address novel candidate genes for INH-induced hepatotoxicity. OBJECTIVE: In this pilot study we analyze a group of 67 cases (who developed hepatotoxicity upon INH treatment) and 111 controls (who did not develop hepatotoxicity) to test for strong genetic effects of NAT2 genotype on the development of toxicity in the Vancouver TB-exposed population. The specific aims of the study were to: 1) determine the spectrum of genetic variation in the NAT2 gene in cases and controls and, 2) to determine whether genetic variants in NAT2 gene correlate with development of hepatotoxicity in the local population. METHODS: 178 patients treated for latent TB at the BC Centre for Disease Control TB Clinic in 2004-05 were enrolled with informed consent. Drug-induced hepatotoxicity was defined by standard criteria. Genomic DNA was isolated from patients’ peripheral lymphocytes. All coding and non-coding exons as well as putative regulatory regions of NAT2 were PCR amplified and bi-directionally sequenced in all participants. RESULTS: To date, 5 of 8 amplicons of the NAT2 gene have been sequenced and 13 variants detected, 5 of which are exonic. Four of the 5 variants in the coding region correspond to amino-acid substitutions and include variants known to encode slow acetylator forms of NAT2. Four of the 8 variants in non-coding exons and putative regulatory regions were previously unreported. Results of the association tests of these variants and their haplotypes, with INH-induced hepatotoxicity, will be presented.