* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PTC Assessment - Teacher Version
Frameshift mutation wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
DNA vaccination wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Koinophilia wikipedia , lookup
Population genetics wikipedia , lookup
Genomic library wikipedia , lookup
Non-coding DNA wikipedia , lookup
DNA supercoil wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Molecular cloning wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genetic drift wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
DNA barcoding wikipedia , lookup
Epigenomics wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Designer baby wikipedia , lookup
History of genetic engineering wikipedia , lookup
SNP genotyping wikipedia , lookup
Metagenomics wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Microsatellite wikipedia , lookup
Point mutation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome editing wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
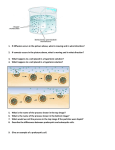
PTC Assessment-Teacher Version ARISE Curriculum. PTC Lab Inquiry-based Assessment S. Gell draft 5 2.2.10 Notes for teachers: The questions are annotated both by level of difficulty and by focus. The difficulty levels roughly correspond with Blooms Taxonomy: Level I. Knowledge, Comprehension Level II Application, Analysis, Level III. Synthesis, Evaluation, Creation. Given that this document is designed to mimic an inquiry-based lab activity, the focus is on the higher levels of Blooms. However, for some students questions that are particularly difficult may not be appropriate. As for content, I have labeled questions that relate directly to the PTC lab as Core Content (CC) while questions that relate to extension activities will be annotated as EX. In the PTC lab we examined natural variation in a human gene for tasting PTC. Use what you learned in this laboratory exercise to help you answer questions about guppies and their ability to see colored patterns. Guppies are small fish that live in warm fresh water. Male guppies are known for their brightly colored patterns. These patterns are thought to be important for mate choice, with females preferring males that have bright and unusual coloring patterns1. ”Sexy” Male Guppy (Image from Ref 1) Q1: (II, CC) A. For a male guppy, what would be one a major advantage and one major disadvantaged of having very brightly colored tails in the wild? [Broad area 1.1] Advantage – brightly colored males would be more attractive to mates and therefore likely to have more offspring. Disadvantage – brightly colored males might be more visible to predators 1 PTC Assessment-Teacher Version Scientists studying guppy behavior noticed that the females needed to be able to detect Ultra-Violet (UV) light to make decisions about the quality of the markings on males2. Humans cannot see colors in this range, but many animals can. In order to detect UV light, animals must have a UV-light receptor in their eye. This receptor gene is called opsin. Scientists sequenced the opsin gene from two related species of guppies. The genomic sequence for part of the UV-light receptor (opsin) for the two guppy species is below. Species 1: P.reticulata CTG GTT TGC TGG ACA CTT TAC GCC AGT GTA GCC TGG TAC ATC TTC TCA AAT CAG _L__V__C__W___T___L__Y__A___S__ V__A___W__Y__ I__ F___S__N__Q Species 2: G. holbrooki CTG GTT TGC TGG ATA CCT TAC GCC AGT GTG GCC TGG TAC ATC TTC ACA CAT CAG _L__V__C__W___T___L__Y__A___S__ V__A___W__Y__ I__ F___I__H__Q Q2: (I,CC) [LS1(9-11)-2a,c] A. Translate each DNA sequence into an amino acid sequence using your codon table. Write the answer on the line provided below the sequence. B. Circle any coding changes in the amino acid sequence. C. Put a box around any silent or non-coding changes in the DNA sequence. Q3: (II or III,CC) A light receptor, like a taste receptor, is used to sense a particular signal and then transmit that information to the brain. How might changes to amino acid sequence effect a light receptor? [LS1(911)FAF+POC-2b] Any reasonable answer would be acceptable. Ideas: Mutations in the receptor could affect its ability to detect light. (ie sensitivity, light wavelength/color) Mutations could also affect the ability of the receptor to send messages into the cell or could affect the receptors ability to be correctly made and placed in the right location in the cell. I would also accept an answer that suggested changes in amino acid sequence 2 PTC Assessment-Teacher Version could change the way the protein folds making it work improperly or differently. Q4: You noticed that sequence TTCTCA (P. reticulata) is recognized by the restriction enzyme FshI, but the sequence to TTCACA in G. holbrooki is not. A. (II, CC) How could you use the restriction enzyme FshI to distinguish between samples of DNA from these two species? I would place both DNA samples (actually PCR products) with the enzyme and only the P. reticulata sample would be cut by the enzyme. When you run the digested samples on a gel the P. reticulata sample would have two bands while the G. holbrooki sample would only have one. Q5. You have discovered a new population of guppies and want to determine if the opsin gene in this population has the same polymorphisms (nucleotide changes) as the other species. A. (II, CC) Explain how the FshI enzyme could be used to help you determine this. If the new population has the P. reticulata allele, the DNA from this population will be cut by the FshI enzyme, if it has the G. holbrooki allele it will not. A1. (III, CC). If the restriction enzyme cuts the DNA from fish in the new population of guppies, can you be confident that the fish carry the P. reticulata allele? If the enzyme does not cut the DNA what does that tell you about the DNA sequence? If FshI cuts the DNA then one can assume that sequence in the new fish is the same as P. reticulata. However, if the new population had a novel gene sequence at this site the enzyme would fail to cut giving a misleading result. *Answering this question correctly requires a high level understanding of the use of restriction enzymes. I would consider it an extension question for students that need an additional challenge. B. (II, CC) Starting with DNA isolation describe the 4 major steps in this experiment and briefly explain why you need to do each. 1. DNA isolation – DNA isolation is required to remove any material (such as DNAases which could degrade DNA) that would interfere with later steps in the experiment. 3 PTC Assessment-Teacher Version 2. PCR amplification – PCR allows us to generate many copies of the sequence of DNA between the primers. This allows us to see the DNA when we run it on a gel 3: Restriction Digest – The restriction digest cuts DNA that carries the sequence recognized by the restriction enzymes. In this experiment it will allow us to determine which allele each fish has. 4. Gel Electrophoresis. Gel electrophoresis will allow us the visualized the digested and undigested DNA and so we can determine the type of alleles in the new population of guppies. C. (II, CC) After you cut the DNA with FshI what would you expect to see if the new population contained fish that were homozygous for the P. reticulata allele? For the G. holbrooki allele? What would you see if there were heterozygous fish? If the population is homozygous for the P. reticulata allele then each sample should have two bands when cut. If the sample is homozygous for G. holbrooki allele then there will only be one band in each sample. If the sample is heterozygous there will be three bands in the digested sample two from the cut P. reticulata allele and one from the uncut G. Holbrooki allele. D. (II,CC) What controls would you include in this experiment to show the genotypes of fish from the new population of guppies? [Broad Area 2.5] Students should include undigested controls for all samples. There should also be samples of both P. reticulata and G. holbrookie (digested and undigested) included as positive controls for comparison. 4 PTC Assessment-Teacher Version Q6: (II, CC) You tested 6 individuals from the new population of guppies. Based on the data below determine if each individual is homozygous for the P. reticulata allele (P/P) or the G. Holbrooki allele (G/G) or is heterozygous for the two alleles (G/P) [Broad area 3.10, LS1(9-11)-2b, LS3(9-11)-7a] U = uncut (not digested with FshI), C = cut (digested with FshI) Genotype of individual samples. (G/G, P/P G/P) 1 __G/G__ 2 __G/P__ 3.___P/P_ 4.___G/P_ 5.___P/P_ 6.___G/P_ Q7: (III, Ex) Following your observation that different populations of guppies have different alleles of the opsin gene, you want to determine which allele represents the ancestral state of the guppy opsin gene. How might you do this? [Broad area 2.4, LS3(9-11)-6a] Compare the sequence of the opsin gene from the guppies to a distantly related species of fish. Whichever species’ opsin sequence most closely matches the sequence of the distantly related species likely represents the ancestral sequence of the gene. 5 PTC Assessment-Teacher Version Q8: (II, Ex) By looking at the data below, determine which allele (P. reticulata or G. holbrooki) is most likely the ancestral state of the guppy opsin and explain how the data supports your hypothesis. [Broad area 1.2 and 4.12, LS3(9-11)-8d] Common ancestor Zebrafish G. holbrooki P. reticulata P.reticulata: TGG ACA CTT TAC GCC AGT GTA GCC TGG TAC ATC TTC TCA AAT CAG G. holbrooki: TGG ATA CCT TAC GCC AGT GTG GCC TGG TAC ATC TTC ACA CAT CAG Zebrafish* TGC ATA CTT TAC GCC AGT GTA GCC TGG TAC ATC TTC TCA AAT CAG Answer here: The sequence from the opsin gene of P. reticulata most closely matches the distantly related species zebrafish. (only one difference in nucleotide sequence, compared to 4 differences for G. holbrookie). Therefore, the common ancestor of both guppies and zebrafish most likely had a sequence close to the P. reticulata sequence. Q9: (III, CC) Imagine that P. reticulata live in only clear, fast moving, rocky streams while G. holbrooki live in slow moving water with lots of green algae that can block the light. Make a hypothesis regarding what selective pressures could be driving the evolution of the opsin gene of these fishes. Remember opsin is important for detection of light in the eye. [LS3(9-11)-7c,8a Broad area 1.1] The mutations in the opsin gene of G. holbrooki may have evolved in response to the selective pressure of low light conditions. Under these conditions the opsin gene may have evolved to be more sensitive to low light or to detect colors of light more likely to penetrate the dark green algae. 6 PTC Assessment-Teacher Version Q10. (III, CC) Based on your data from the newly discovered population of guppies, what type of environment do you think these fish might have come from? Explain your reasoning. [Broad area 1.1, 4.12] I would accept any reasonable answer if well supported. Here are a few examples. 1. The new population might live in a mixed environment where they experience both clear quick water and slow green water either because of water quality changes at different times of year or because of the close proximity of these different environments. This is suggested by the presence of both allele types. If different allele types confer different abilities to see light, fish with both receptors might have an advantage in a mixed environment. 2. The fish may have recently shifted from one type of environment to another and are currently undergoing adaptation and population shift from mostly one allele type to another. 3. There are slightly more P alleles (7) in our sample population than G alleles (5), this might lead students to conclude the water is more like the environment for P. reticulata than the environment of G. holbrooki. References 1. Olendorf et al. Frequency-Dependent Survival in Natural Guppy Populations. Nature. Vo1 144. 1 June 2006. 2. Smith et al. Ultraviolet Vision and Mate Choice in the Guppy. Behavioral Ecology. Vo1 13. 2002. * Not based on real data. 7