* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download AP Bio Ch 4
Survey
Document related concepts
Cytoplasmic streaming wikipedia , lookup
Cellular differentiation wikipedia , lookup
Cell growth wikipedia , lookup
Cell culture wikipedia , lookup
Extracellular matrix wikipedia , lookup
Cell encapsulation wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Cell nucleus wikipedia , lookup
Signal transduction wikipedia , lookup
Cell membrane wikipedia , lookup
Cytokinesis wikipedia , lookup
Transcript
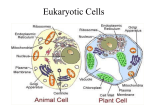
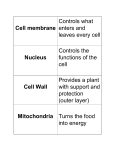
Chapter 7 A Tour of the Cell cell - basic unit of structure and function in all organisms The cell demonstrates 4 themes: emergent properties correlation of structure and function interactions of organisms within their environment evolution Robert Hooke - first described cells using a light microscope (1665) - coined the term “cell” light microscope - condenser lens focuses light on a specimen - light passing through the specimen is refracted (bent) with an objective lens and an ocular lens - image is thus magnified and inverted magnification - how much larger an object is made to appear compared to its real size - highest magnification of a light microscope with maximum resolution is about 1000 times resolution - minimum distance between 2 points that can still be distinguished as 2 separate points (clarity) - maximum possible resolution of a light microscope is 0.2 μm electron microscope - far surpassed the resolving power of the light microscope - uses a beam of electrons - practical resolution of about 2 nm - can only view dead specimens 2 types of electron microscopes: transmission electron microscope (TEM) - aims a beam of electrons at a thin section of specimen electrons transmitted through the specimen are focused and the image magnified by using electromagnetic lenses to bend the paths of the electrons - image is focused onto a viewing screen or film - used to study internal cell structures scanning electron microscope (SEM) - useful for studying the surface of a specimen - electron beam scans the surface of a specimen - this excites secondary electrons on the sample’s surface which are collected and focused on a viewing screen cytology - study of cell structure cell fractionation - centrifuging disrupted cells to isolate components of various sizes, densities, and shapes - uses an ultracentrifuge (spins up to 80,000 rpm) steps of cell fractionation: homogenization of tissue using pistons, blenders, or ultrasound slow centrifugation of homogenate nuclei and other larger particles settle to bottom of tube forming a pellet unpelleted fluid (supernatant) decanted into another tube supernatant centrifuged at a faster speed separating out smaller organelles supernatant decanted again and centrifuged at higher speed (and so on) 2 major kinds of cells: prokaryotic and eukaryotic prokaryotic eukaryotic bacteria, archaebacteria no true nucleus; lacks nuclear envelope genetic material in nucleoid region no membrane-bound organelles protists, fungi, plants, animals true nucleus; bounded by nuclear envelope nuclear material in nucleus cytoplasm (cytosol and membrane-bound organelles) 10 – 100 μm 0.1 – 10 μm cytoplasm - region between cell membrane and nucleus cytosol - semi-fluid part of cytoplasm Range of cell size limited by metabolic requirements. lower limits: probably determined by the smallest size with enough DNA to program metabolism and enough ribosomes, enzymes, etc. to sustain life upper limits: determined by the surface area to volume ratio (as a cell increases in size, its volume grows proportionately more than its surface area) - surface area must be large enough for cell volume (to provide adequate exchange surface for oxygen, nutrients, and wastes) plasma membrane - boundary of every cell - selective barrier - allows passage of materials into and out of cell internal membranes in eukaryotic cells: separate cell into compartments have unique lipid and protein compositions may take part in metabolic reactions (many enzymes incorporated into membranes) provide localized environmental conditions necessary for specific metabolic processes isolate reactions Nucleus nucleus - membrane-bound organelle in a eukaryotic cell - contains most of the genes that control the entire cell - about 5 μm in diameter - enclosed by nuclear envelope nuclear envelope - double membrane enclosing the nucleus - 2 lipid bilayer membranes separated by a space - contains pores (regulate passage of molecules into and out of nucleus) nuclear lamina - network of protein filaments on envelope’s nuclear side - stabilizes nuclear shape chromatin - genetic material (complex of DNA and histone proteins) - make up chromosomes in eukaryotic cells chromosomes - long, threadlike group of genes - composed of chromatin - each species has a characteristic number of chromosomes - human cells have 46 chromosomes (except sperm and egg cells) nucleolus - spherical region in nucleus of nondividing cells - may be 2 or more - make ribosomal subunits from ribosomal RNA and proteins - subunits pass through pores to cytoplasm where they are assembled protein synthesis - controlled by nucleus - messenger RNA (mRNA) transcribed in nucleus from DNA - mRNA passes through pores into cytoplasm - mRNA attaches to ribosomes where “message” is translated into a protein Ribosomes ribosome - cytoplasmic organelle - site of protein synthesis - consist of RNA and protein - made in nucleolus (in eukaryotic cells) - many are found in cells with high rates of protein synthesis - found free in cytoplasm or bound to endoplasmic reticulum free ribosomes - in cytosol - most proteins made here function in cytosol bound ribosomes - attached to outside of ER - usually make proteins for membrane inclusion or export - many found in cells specializing in protein excretion (pancreas) endomembrane system vesicles - membranes vary in structure and function - may change in composition, thickness, and behavior - includes: nuclear envelope ER Golgi apparatus lysosomes vacuoles plasma membrane - membrane-enclosed sacs that are pinched off portions of membranes moving from 1 membrane to another Endoplasmic Reticulum endoplasmic reticulum cisternal space 2 regions of ER smooth ER rough ER - membranous network of tubules and sacs (cisternae) - most extensive portion of endomembranous system - continuous with external membrane of nuclear envelope - internal lumen of ER - continuous with space between membranes of nuclear envelope - cytoplasmic surface lacks ribosomes - cytoplasmic surface studded with ribosomes - continuous with outer membrane of nuclear envelope functions of smooth ER: participates in synthesis of lipids, phospholipids, & steroids - example: mammalian sex hormones - cells that secrete these products are rich in smooth ER (testes, ovaries, skin oil glands) participates in carbohydrate metabolism - smooth ER in liver contains an enzyme that helps convert glycogen to glucose detoxifies drugs and poisons - smooth ER in liver contains enzymes which detoxify drugs and poisons - enzymes catalyze addition of a hydroxyl group to drugs and poisons they become soluble in cytosol can be excreted - smooth ER in liver cells increases in response to barbiturates, alcohol and other drugs (may increase drug tolerance) store calcium ions necessary for muscle contraction - in muscle cells, ER pumps Ca2+ from cytosol into cisternal space - in response to nerve impulse, Ca2+ leaks back into cytosol which triggers cell contraction functions of rough ER manufactures secretory proteins membrane production rough ER and protein synthesis - ER ribosomes synthesize secretory proteins - growing polypeptide is threaded through ER membrane into cisternal space - protein folds into its native configuration - if destined to be a glycoprotein, enzymes catalyze the bonding of an oligosaccharide to the protein - transport vesicle containing the protein pinches off from a specialized region of ER (transitional ER) glycoproteins oligosaccharide transport vesicle - protein covalently bonded to carbohydrate - small polymer of sugar units - membrane vesicle in transport from 1 part of the cell to another rough ER and membrane production - membrane proteins produced by ER ribosomes (polypeptide is inserted into rough ER membrane) - enzymes within ER membrane synthesize phospholipids from materials in cytosol - proteins and phospholipids are assembled and expand ER membrane - expanded membrane transported as a vesicle to other parts of the cell Golgi Apparatus Golgi apparatus - organelle made of flattened membranous sacs (cisternae) - many transport vesicles leave ER and travel to Golgi - finishes, stores, and ships ER products - has 2 poles: cis face and trans face cis face - “forming face” of Golgi - accepts transport vesicles from nearby transitional ER - vesicle fuses its membrane with the membrane of the cis face and empties its contents into Golgi cisternal space trans face - “maturing face” of Golgi - vesicles pinch off from Golgi here to transport molecules to other sites Golgi - enzymes in cisternae modify ER products as they move from cis to trans face - each cisterna has unique enzymes - products move from cisterna to cisterna in transport vesicles some examples of what the Golgi does to ER products: alters some membrane phospholipids alters oligosaccharide portion of glycoproteins tags some products to be sent to various cell parts (phosphate groups or oligosaccharides may be added to act as molecular identification tags) sorts products for secretion (these products leave trans face in transport vesicles which fuse with plasma membrane) Lysosomes lysosomes - membrane-enclosed bag of hydrolytic enzymes - enzymes digest macromolecules - probably pinch off from trans face of Golgi lysosomal enzymes - lipases, carbohydrases, proteases, nucleases - optimal pH 5 - made in RER; processed in Golgi lysosomal membrane - keeps potentially destructive enzymes separate from cytosol - pumps H+’s inward to keep optimal pH for enzymes - made in RER; processed in Golgi functions of lysosomes intracellular digestion - lysosomes fuse with food-filled vacuoles - hydrolytic enzymes digest food - example: amoeba eating by phagocytosis - example: human cells called macrophages “eat” bacteria by phagoctosis phagocytosis - plasma membrane engulfs substances and pinches off to form a particlecontaining vacuole recycling of cell parts - lysosomes engulf other organelles or part of cytosol and digest them - monomers released into cytosol where they can be recycled programmed cell destruction - important during metamorphosis and development lysosomal storage diseases - inherited diseases resulting from impaired lysosomal function - lack of specific lysosomal enzymes cause some substances to accumulate interferes with cell function - examples: Pompe’s disease - a carbohydrase that breaks down glycogen is missing - glycogen accumulates and damages liver Tay-Sach’s Disease - a lipase is missing - lipids accumulate in brain Vacuoles vacuole - membrane-enclosed organelle - larger than a vesicle food vacuole - formed by phagocytosis - site of intracellular digestion in some protists and macrophages contractile vacuole - pumps excess water from cell - found in some freshwater protozoa central vacuole - large vacuole found in most mature plant cells - forms when smaller vacuoles from ER and Golgi coalesce - enclosed by tonoplast (part of endomembrane system) - functions: stores organic compounds (protein storage in seeds, etc.) stores inorganic ions (K+, Cl-) sequesters dangerous metabolic byproducts from cytoplasm contains pigments in some cells (pigments in flowers) may contain poisonous or unpalatable compounds for protection against predators absorbs water and elongates cell (promotes plant growth) contributes to large ratio of membrane surface area to cytoplasmic volume (only thin layer of cytoplasm between tonoplast and plasma membrane) relationships among endomembranes: Nucl ear IS A N E X T E N S IO N O F envel ope IS C O N F L U E N T W IT H Rough ER s moot h er MEM BRANE AND SECR ETO RY P R O T E IN S P R O D U C E D IN E R A R E T R A N S P O R T E D IN ves i cl es F U S E W IT H T H E F O R M IN G F A C E O F gol gi appar at us P IN C H E S O F F M A T U R IN G F A C E G IV E R IS E T O ves i cl es Peroxisomes F U S E W IT H A N D A D D T O PLASMA M EM BR A NE AN D M A Y RE LE AS E C E LLU LAR P R O D U C T S T O O U T S ID E Lys os omes PLASMA & VACUOLES MEMBRANE peroxisomes - membrane-bound organelles that contain specialized enzymes - found in nearly all eukaryotic cells - probably form by pinching off from existing peroxisomes - contain peroxide-producing oxidases transfer hydrogen to oxygen producing hydrogen peroxide oxidase RH + O -------- R + H O 2 2 > 2 2 - contain catalase converts hydrogen peroxide to water catalase 2H2O2 ------->2H2O +O2 functions of peroxisomal reactions break down of fatty acids into smaller molecules (acetyl coA) detoxification of alcohol and poisons glyoxysomes - specialized peroxisomes found in germinating seeds - contain enzymes that convert lipids to carbohydrates - make energy stored in seed oils available for the seedling mitochondria and chloroplasts mitochondria and chloroplasts - organelles that change energy from surrounding into useable forms of energy - enclosed by double membranes - not part of endomembrane system (membrane proteins made by free ribosomes and by ribosomes in these organelles themselves) - contain ribosomes and some DNA that programs some of their protein synthesis (most programmed by nuclear DNA and synthesized in cytosol) - semiautonomous grow & reproduce within cell endosymbiotic theory - says that mitochondria and chloroplasts originated when a eukaryotic cell engulfed a prokaryotic cell - mitochondria came about when a eukaryotic cell engulfed an oxygenusing non-photosynthetic prokaryote - chloroplasts came about when a eukaryotic cell engulfed a photosynthetic prokaryote evidence: both mitochondria and chloroplasts have 2 membranes surrounding them both contain ribosomes and their own DNA both are autonomous (grow & reproduce within the cell) mitochondria respiration - catabolic, oxygen-requiring process that uses energy extracted from organic molecules to produce ATP mitochondria - sites of cellular respiration - found in nearly all eukaryotic cells - number per cell correlates with the cell’s metabolic activity - dynamic structures (move, change shape, divide) mitochondrion structure - enclosed by 2 phospholipid bilayers with unique proteins embedded in them - outer membrane – smooth - highly permeable to small solutes - blocks passage of proteins and other macromolecules - inner membrane - contains cristae (membrane infoldings) - contains embedded enzymes involved in respiration - cristae increase surface area for respiration reactions - 2 internal compartments (separated by internal membrane) intermembrane space - narrow region between inner and outer membranes - reflects solute composition of cytosol mitochondrial matrix - enclosed by inner mitochondrial membrane - contains enzymes that catalyze many steps of respiration chloroplasts plastids - membrane-bound organelles in plants and algae - contain amyloplasts, chromoplasts, and chloroplasts amyloplasts - colorless plastids - store starch - found in roots and tubers chromoplasts - pigment-containing plastids (other than chlorophyll) - responsible for colors of fruits, flowers, and autumn leaves chloroplasts - chlorophyll-containing plastids - bound by a double membrane - sites of photosynthesis - found in eukaryotic algae, leaves, and other green plant parts - dynamic structures (move, change shape, divide) chloroplast structure: - contain thylakoids (flattened membranous sacs) - membranes contain chlorophyll - take part in initial steps of photosynthesis - some are stacked into grana - 3 compartments formed by membranes intermembrane space - narrow space between the 2 membranes surrounding the chloroplast thylakoid space - space inside a thylakoid stroma - space between thylakoids and inner membrane - contains viscous fluid - site of photosynthetic reactions that convert CO2 to sugar Cytoskeleton cytoskeleton - network of fibers throughout the cytoplasm - forms a dynamic framework for support, movement, and regulation - supports cell; helps maintain cell shape - enables a cell to change shape - takes part in motility interacts with proteins called motor molecules (e. g. organelle movement, muscle contraction, locomotor organelles) - takes part in regulation transmits signals from cell’s surface to its interior - consists of at least 3 types of fibers: microtubules (thickest) microfilaments (thinnest) intermediate filaments microtubules - found in cytoplasm of all eukaryotic cells - structure: straight hollow fibers constructed from a globular protein called tubulin - each tubulin molecule consists of 2 polypeptide subunits (α-tubulin and β-tubulin) - may be disassembled and recycled elsewhere in the cell - functions: cell support; reinforce cell shape tracks for organelle movement (protein motor molecules interact with microtubules to translocate organelles (e.g. vesicles from Golgi to plasma membrane) separation of chromosomes during cell division centrioles - pair of cylindrical structures located in the centrosome in animal cells - 9 sets of triplet microtubules arranged in a ring - at right angles to each other - replicate during cell division - may organize microtubule assembly during cell division cilia & flagella- locomotor organelles found in some eukaryotic cells - made of microtubules - propel many eukaryotic unicellular organisms through water - flagella propel motile sperm cells (animals, algae, some plants) - cilia draw fluid across the surface of stationary cells (ciliated cells of trachea) structure of cilia and flagella - extensions of plasma membrane with a core of microtubules - “9 + 2 pattern” - core made of 9 microtubule doublets in a ring around 2 single microtubules in center (doublet = pair of attached microtubules) - radial spokes - connect each doublet to the center - side arms - attach each doublet to neighboring doublet - made of protein dynein - basal bodies - anchor the microtubular assembly of cilia & flagella - structurally identical to centrioles (can convert into a centriole or vice versa) dynein - motor molecule that changes its shape using ATP as energy - shape change and the resistance of the radial spokes causes cilia or flagella to bend microfilaments - actin filaments - structure: consist of 2 long actin chains wound into a helix each chain is made of globular protein monomers called G-actin - functions: provide cellular support - form a network with other proteins just inside plasma membrane muscle contraction - interact with thicker filaments made of the protein myosin in muscle cells localized contraction of cells - small actin-myosin aggregates in some parts of cell cause localized contractions (e.g. pinching an animal cell in 2 during cell division; elongation and contraction of pseudopodia during amoeboid movement) cytoplasmic streaming in plant cells - circular flow of cytoplasm (to distribute materials may involve actin-myosin interactions) intermediate filaments - different in diameter and composition depending on cell type - structure: made of keratin subunits more permanent than microtubules and microfilaments - functions: bear tension (may serve as framework for cytoskeleton) reinforce cell shape may fix organelles in place (e.g. nucleus) make up nuclear lamina Cell Surfaces and Junctions cell wall - external to plasma membrane in plant cells - thicker than plasma membrane - structure: chemical composition differs from cell to cell and species to species consists of strong cellulose fibers in a matrix of other polysaccharides and proteins - functions: protects plant cell maintains cell shape prevents excess water uptake plasmodesmata - channels through cell walls connecting cytoplasm of neighboring cells development of plant cell walls: young cell secretes thin primary cell wall middle lamella (made of sticky polysaccharides called pectin) between adjacent primary cell walls cements cells together cell stops growing cells secretes hardening substances into primary cells wall secondary cell wall added between plasma membrane and primary cell wall secondary cell wall - durable matrix - supports and protects cell extracellular matrix (ECM) of animal cells ECM - meshwork of macromolecules outside plasma membrane - secreted by cells - composed mostly of glycoproteins (mostly collagen) collagen - about ½ total protein in vertebrates - forms strong extracellular fibers embedded in a meshwork of glycoproteins called proteoglycans - some cells are directly attached to the ECM; others are attached to ECM by glycoproteins called fibronectins - functions: supports and anchors cells helps control gene activity (chemical signals in ECM may trigger specific gene transcription) intercellular junctions plants: plasmodesmata - channels that perforate plant cell walls connecting cytoplasm of adjacent cells - lined by plasma membrane - allows passage of water and small solutes (enhanced by cytoplasmic streaming) animals: tight junctions - hold cells together tightly enough to block transport of substances through the intercellular space - specialized membrane proteins in adjacent cells bond to each other - usually occur in belts around each cell (may separate 2 kinds of solutions) desmosomes - rivet cells together into strong sheets - permit substances to pass freely through intracellular spaces - made of: glycoprotein filaments that penetrate and attach plasma membranes of both cells a dense disk inside plasma membrane reinforced by intermediate filaments (keratin) gap junctions - specialized for moving materials between the cytoplasm of adjacent cells - formed by 2 connecting protein rings each embedded in plasma membranes of adjacent cells - form pores big enough to allow cells to share smaller molecules (ions, sugars, amino acids, vitamins) but not macromolecules (like proteins) - common in animal embryos and cardiac muscle (chemical communication between cells is essential)