* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Types of plasmid One way of grouping plasmids is by their ability to
Genealogical DNA test wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Minimal genome wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Primary transcript wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Non-coding DNA wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Epigenomics wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Point mutation wikipedia , lookup
DNA supercoil wikipedia , lookup
Genome editing wikipedia , lookup
Designer baby wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Molecular cloning wikipedia , lookup
Microevolution wikipedia , lookup
Genetic engineering wikipedia , lookup
Helitron (biology) wikipedia , lookup
Genomic library wikipedia , lookup
DNA vaccination wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
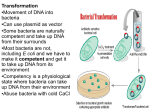
Types of plasmid One way of grouping plasmids is by their ability to transfer to other bacteria. Conjugative plasmids contain so-called tra-genes, which perform the complex process of conjugation, the sexual transfer of plasmids to another bacterium (Fig. 4). Non-conjugative plasmids are incapable of initiating conjugation, hence they can only be transferred with the assistance of conjugative plasmids, by 'accident'. An intermediate class of plasmids are mobilisable, and carry only a subset of the genes required for transfer. These plasmids can 'parasitise' another plasmid, transferring at high frequency in the presence of a conjugative plasmid. Figure 4 : Schematic drawing of bacterial conjugation. 1 Chromosomal DNA. 2 Plasmids. 3 Pilus. 1 It is possible for several different types of plasmids to coexist in a single cell, e.g., seven different plasmids have been found in E. coli. On the other hand, related plasmids are often 'incompatible', resulting in the loss of one of them from the cell line. Therefore, plasmids can be assigned into incompatibility groups, depending on their ability to coexist in a single cell. These incompatibility groupings are due to the regulation of vital plasmid functions. An obvious way of classifying plasmids is by function. There are five main classes: Fertility-(F)plasmids, which contain tra-genes. They are capable of conjugation. Resistance-(R)plasmids, which contain genes that can build a resistance against antibiotics or poisons. Historically known as Rfactors, before the nature of plasmids was understood. Col-plasmids, which contain genes that code for (determine the production of) colicines, proteins that can kill other bacteria. Degrative plasmids, which enable the digestion of unusual substances, e.g., toluene or salicylic acid. Virulence plasmids, which turn the bacterium into a pathogen. Plasmids can belong to more than one of these functional groups. Plasmids that exist only as one or a few copies in each bacterium are, upon cell division, in danger of being lost in one of the segregating bacteria. Such single-copy plasmids have systems which attempt to actively distribute a copy to both daughter cells. Some plasmids include an addiction system. These plasmids produce both a long-lived poison and a short-lived antidote. Daughter cells that retain a copy of the plasmid survive, while a daughter cell that fails to inherit the plasmid dies or suffers a reduced growth-rate because of the lingering poison from the parent cell. This is an example of plasmids as selfish DNA. 2 Applications of plasmids Plasmids serve as important tools in genetics and biochemistry labs, where they are commonly used to multiply (make many copies of) or express particular genes. There are many plasmids that are commercially available for such uses. Initially, the gene to be replicated is inserted in a plasmid. These plasmids contain, in addition to the inserted gene, one or more genes capable of providing antibiotic resistance to the bacterium that harbors them. The plasmids are next inserted into bacteria by a process called transformation, which are then grown on specific antibiotic(s). Bacteria which took up one or more copies of the plasmid then express (make protein from) the gene that confers antibiotic resistance. This is typically a protein which can break down any antibiotics that would otherwise kill the cell. As a result, only the bacteria with antibiotic resistance can survive, the very same bacteria containing the genes to be replicated. The antibiotic(s) will, however, kill those bacteria that did not receive a plasmid, because they have no antibiotic resistance genes. In this way the antibiotic(s) acts as a filter selecting out only the modified bacteria. Now these bacteria can be grown in large amounts, harvested and lysed to isolate the plasmid of interest. Another major use of plasmids is to make large amounts of proteins. In this case you grow the bacteria containing a plasmid harboring the gene of interest. Just as the bacteria produces proteins to confer its antibiotic resistance, it can also be induced to produce large amounts of proteins from the inserted gene. This is a cheap and easy way of mass-producing a gene or the protein it then codes for--for example, insulin or even antibiotics. Plasmid Sex .....In most bacteria there are several pieces of DNA. .. One is the somatic genome - a huge circle of double-stranded DNA that actually measures about 2 mm in length, and is all crammed into the little cell. ..This large piece of DNA is what defines the type of bacterium it is. .. The cell cannot live without this circle of DNA. In addition, there are various optional smaller circles of DNA, which are usually called plasmids... To repeat: ..these are 'optional' and the cell can get along without them unless the genes on those plasmids allow it to survive under unusual conditions such as when a particular antibiotic is in the neighborhood, and the plasmid contains a 3 gene that protects the cell from that antibiotic by any one of several different mechanisms. .....A cell can duplicate a plasmid and then send one copy over to another cell via a thin tube called a pilus. ..Seemingly, each type of plasmid codes for its own special type of pilus. .....A widely used plasmid of E. coli is one called "F" (for fertility). .. Cells that possess "F" are called male (F-donors or "F+"), these cells usually possess two F-pili for the transport of the F into cells lacking it. .. Those without F are called females (potential recipients, F-). .. In this exercise you will mix a very few F+ with a much larger number of F-, and show that the Fcells are converted to F+ at a rather rapid pace. ..The rapidity is because because it is a chain-reaction as the newly form F+ cells are added to the pool of donors. ..(You can change this exercise into an experiment if you can design an experiment from which you can determine the time it takes to convert an F- into an F+.) .....First, you must have a strain of donor that possesses a somatic genome that is different from that of the recipient. ..That way you can "select against" the original donor cells by using a mating medium in which they cannot grow, but in which the recipients can. ..For this we will use donors that are normal with respect to sensitivity to the antibiotic streptomycin, while the recipients will be streptomycin resistant (str R). ..We will add streptomycin to the medium, and none of the original donors will be able to grow up to form colonies. ..(Always keep in the back of your mind a very important word: "controls"!) .....Second, you must have a way to tell whether or not the recipients have acquired the F. ..We will do a little trick here, and use an F that has the lactose operon inserted into it. ..This is called "F-lac." ..(Whenever other genes are incorporated into F, the F is now called a type of F-prime (F'). ..Thus F-lac is a type of F'. ..We will therefore start with F- cells that do not have a functional lac-operon (lac-), and so when they acquire the F-lac, they will be able to use lactose sugar to grow. .. (This is not a genetically engineered product, but one that can be made naturally, and therefore doesn't not need to be approved by an institutional recombinant DNA committee. ..Indeed, no part of this exercise is considered genetic engineering. ..It is all genetic recombination using natural means. ..That is one of the important 4 things about this exercise - this happens naturally around you all the time in the genetically mobile and fluid microbial world.) Figure 2: Transfer of Genetic Material through transformation, translation, and conjugation figure taken from Levy, 1998 see reference in text Conjugation Conjugation plays a large role in the spread of antibiotic resistance through bacteria. This process involves direct cell-to-cell contact of two bacterial cells, and the subsequent transfer of DNA. Conjugation can occur between species that are unrelated; for this reason, a large gene pool is available from which bacteria can exchange and acquire new genetic material. (Guiney, 1984). Sex pili make contact between the donor and the recipient cell. Once the two cell walls are in contact, this allows a mating bridge to form. The plasmid DNA in the donor, possibly containing antibiotic resistance genes, is nicked in one strand; this strand proceeds into the recipient cell by undergoing rolling-circle replication (Hartl and Jones, 5 1998, p316). Figure 3 demonstrates the process of rolling-circle replication. Complementary copies of the DNA are produced in both the donor and the recipient cells. Finally, the linear plasmid in the recipient becomes circular and is ligated, and then both of the cells have a copy of the plasmid. B. Conjugation - pilus transfers plasmid from one bacteria to another 6 Figure 3: Rolling Circle Replication There are certain barriers to the process of conjugation, which often play a role in the evolution of resistance. First, interactions at the cell surface are involved. Contact must occur for mating to take place. Next, foreign DNA is susceptible to host restriction modification systems which target and cleave foreign DNA. Third, the new plasmid be unable to replicate in the new host. An appropriate origin of replication in the new host may not be available, thus preventing replication. For example, transferred DNA is more likely to be stable in a new host if it contains fewer restriction enzyme sites. This makes it less likely that the DNA will be degraded by restriction enzymes which attack foreign DNA. A selective advantage exists for plasmids with fewer restriction sites. Selective 7 advantages which aid in stability can parallel the beginnings of the evolution of strains that are resistant to antibiotics. Antibiotic resistance will spread more readily if the resistance genes can be transferred into non-resistant cells. The ability to both invade many types of cells and to be maintained in these cells are traits that are advantageous for survival and transfer of plasmids. 8 9 Transduction Transduction occurs when a bacteriophage carries DNA from one species to another. When a bacteriophage destroys its current host and invades a new one, it may carry pieces of chromosomal DNA or plasmids from the previous host. An occasional phage may carry some bacterial DNA. Recombination can then occur between the phage (carrying bacterial DNA) and the new host's bacterial DNA. Transfer of DNA thus occurs from one bacterial cell to another, carried by a bacteriophage. ( Lacey, 1984 and Hartl and Jones, 1998, p308). This provides another method for the spread of resistance among bacteria. 10 D. Transduction - a virus infects to bacteria carrying genetic material form one bacteria to another 11 What kind of genetic variation does this represent 12 Transformation Transformation is another method of acquiring resistance. During transformation, bacterial cells take up DNA from the surrounding environment. Certain requirements exist in order for transformation to take place. First, exogenous DNA must be present in the immediate environment. Bacteria must have mechanisms that allow the DNA to be taken up through the bacterial cell walls. Also, the DNA must be incorporated into the chromosome of the host, often by homologous recombination. During homologous recombination, parts of the chromosome are replaced with 13 related DNA (Maiden, 1998). Restriction modification systems play a role in transformation as well as in conjugation. However, it is thought that since these modification systems generate both DNA ends and smaller fragments, restriction modification may actually increase the chance of recombination with incorporated fragments. This could occur because recombination occurs more frequently if the ends are homologous. Transforming E. coli Treatment of E. coli with the mixture of religated molecules will produce some colonies that are able to grow in the presence of both ampicillin and kanamycin. A suspension of E. coli is treated with the mixture of religated DNA molecules. The suspension is spread on the surface of agar containing both ampicillin and kanamycin. The next day, a few cells — resistant to both antibiotics — will have grown into visible colonies containing billions of transformed cells. Each colony represents a clone of transformed cells. However, E. coli can be simultaneously transformed by more than one plasmid, so we must demonstrate that the transformed cells have acquired the recombinant plasmid. Electrophoresis of the DNA from doubly-resistant colonies (clones) tells the story. Plasmid DNA from cells that acquired their resistance from a recombinant plasmid only show only the 3755-bp and 1875-bp bands (Clone 1, lane 3). 14 Clone 2 (Lane 4) was simultaneous transformed by religated pAMP and pKAN. (We cannot tell if it took up the recombinant molecule as well.) Clone 3 (Lane 5) was transformed by the recombinant molecule as well as by an intact pKAN. C. Transformation - Pieces of DNA are absorbed into a competent cell 15 What does this tell us about the virulence factor added by the capsule? Transformation with the Jellyfish Gene Cloning other Genes The recombinant vector described above could itself be a useful tool for cloning other genes. Let us assume that within its kanamycin resistance gene (kanr) there is a single occurrence of the sequence 5'GAATTC 3' 3' CTTAAG 5' This is cut by the restriction enzyme EcoRI, producing sticky ends. If we treat any other sample of DNA, e.g., from human cells, with EcoRI, fragments with the same sticky ends will be formed. Mixed with EcoRItreated plasmid and DNA ligase, a small number of the human molecules 16 will become incorporated into the plasmid which can then be used to transform E. coli. But how to detect those clones of E. coli that have been transformed by a plasmid carrying a piece of human DNA? The key is that the EcoRI site is within the kanr gene, so when a piece of human DNA is inserted there, the gene's function is destroyed. All E. coli cells transformed by the vector, whether it carries human DNA or not, can grow in the presence of ampicillin. But E. coli cells transformed by a plasmid carrying human DNA will be unable to grow in the presence of kanamycin. So, Spread a suspension of treated E. coli on agar containing ampicillin only grow overnight with a sterile toothpick transfer a small amount of each colony to an identified spot on agar containing kanamycin (do the same with another ampicillin plate) Incubate overnight All those clones that continue to grow on ampicillin but fail to grow on kanamycin (here, clones 2, 5, and 8) have been transformed with a piece of human DNA. Some recombinant DNA products being used in human therapy Using procedures like this, many human genes have been cloned in E. coli or in yeast. This has made it possible — for the first time — to produce unlimited amounts of human proteins in vitro. Cultured cells (E. coli, yeast, mammalian cells) transformed with the human gene are being used to manufacture: 17 insulin for diabetics factor VIII for males suffering from hemophilia A factor IX for hemophilia B human growth hormone (GH) erythropoietin (EPO) for treating anemia three types of interferons several interleukins granulocyte-macrophage colony-stimulating factor (GM-CSF) for stimulating the bone marrow after a bone marrow transplant granulocyte colony-stimulating factor (G-CSF) for stimulating neutrophil production, e.g., after chemotherapy and for mobilizing hematopoietic stem cells from the bone marrow into the blood. tissue plasminogen activator (TPA) for dissolving blood clots adenosine deaminase (ADA) for treating some forms of severe combined immunodeficiency (SCID) angiostatin and endostatin for trials as anti-cancer drugs parathyroid hormone leptin hepatitis B surface antigen (HBsAg) to vaccinate against the hepatitis B virus Bacterial Transformation This is a very basic technique that is used on a daily basis in a molecular biological laboratory. The purpose of this technique is to introduce a foreign plasmid into a bacteria and to use that bacteria to amplify the plasmid in order to make large quantities of it. This is based on the natural function of a plasmid: to transfer genetic information vital to the survival of the bacteria. Bacterial Transformation and Transfection Bacterial transformation is the process by which bacterial cells take up naked DNA molecules. If the foreign DNA has an origin of replication recognized by the host cell DNA polymerases, the bacteria will replicate the foreign DNA along with their own DNA. When transformation is coupled with antibiotic selection techniques, bacteria can be induced to uptake certain DNA molecules, and those bacteria can be selected for that incorporation. Bacteria which are able to uptake DNA are called 18 "competent" and are made so by treatment with calcium chloride in the early log phase of growth. The bacterial cell membrane is permeable to chloride ions, but is non-permeable to calcium ions. As the chloride ions enter the cell, water molecules accompany the charged particle. This influx of water causes the cells to swell and is necessary for the uptake of DNA. The exact mechanism of this uptake is unknown. It is known, however, that the calcium chloride treatment be followed by heat. When E. coli are subjected to 42degC heat, a set of genes are expressed which aid the bacteria in surviving at such temperatures. This set of genes are called the heat shock genes. The heat shock step is necessary for the uptake of DNA. At temperatures above 42degC, the bacteria's ability to uptake DNA becomes reduced, and at extreme temperatures the bacteria will die. A Model of a Bacterial Plasmid Legend: Plasmids are similar to viruses, but lack a protein coat and cannot move from cell to cell in the same fashion as a virus. Plasmid vectors are small circular molecules of double stranded DNA 19 derived from natural plasmids that occur in bacterial cells. A piece of DNA can be inserted into a plasmid if both the circular plasmid and the source of DNA have recognition sites for the same restriction endonuclease. The plasmid and the foreign DNA are cut by this restriction endonuclease (EcoRI in this example) producing intermediates with sticky and complementary ends. Those two intermediates recombine by base-pairing and are linked by the action of DNA ligase. A new plasmid containing the foreign DNA as an insert is obtained. A few mismatches occur, producing an undesirable recombinant. The new plasmid can be introduced into bacterial cells that can produce many copies of the inserted DNA . This technique is called DNA cloning. The Recipe for Successful Interactions First, it is necessary to construct the 'bait' and 'hunter' fusion proteins. The 'bait' fusion protein is the protein of interest (or 'bait') linked to the GAL4 binding domain, or GAL4 BD. This is done by inserting the segment of DNA encoding the bait into a plasmid, which is a small circular molecule of double-stranded DNA that occurs naturally in both bacteria and yeast. This plasmid will also have inserted in it a segment of Gal4 BD DNA next to the site of bait DNA insertion. Therefore, when the DNA from the plasmid is transcribed and converted to protein, the bait will now have a binding domain attached to its end (Figure 3). The same procedure is used to construct the 'hunter' protein, where the potential binding partner is fused to the GAL4 AD. In addition to having the fusion proteins encoded for, these plasmids will also contain selection genes, or genes encoding proteins that contribute to a cell's survival in a particular environment. An example of a selection gene is one encoding antibiotic resistance; when antibiotics are introduced, only cells with the antibiotic resistance gene will survive. Yeast two-hybrid assays typically use selection genes encoding proteins capable of synthesizing amino acids such as histidine, leucine and tryptophan (Figure 4). 20 Figure 4. Bait and Hunter Plasmids. The yeast two-hybrid assay uses two plasmid constructs: the bait plasmid, which is the protein of interest fused to a GAL4 binding domain, and the hunter plasmid, which is the potential binding partner fused to a GAL4 activation domain. Once the plasmids have been constructed, they must next be introduced into a host yeast cell by a process called "transfection". In this process, the outermembrane of a yeast cell is disturbed by a physical method, such as sonification or chemical disruption. This disruption produces holes that are large enough for the plasmid to enter, and in this way, the plasmids can cross the membrane and enter the cell (Figure 5). Once the cells have been transfected, it is necessary to isolate colonies that have both 'bait' and 'hunter' plasmids. This is because not every cell will have both plasmids cross their plasma membrane; some will have only one plasmid, while others will have none. Isolation of transfected cells involves identifying cells containing plasmids by virtue of their expressing the selection genes mentioned previously. After the cells have been transfected and allowed to recover for several days, they are then plated on minimal media, or media that is lacking one essential nutrient, such as tryptophan. The cells used for transfection are called auxotrophic mutants; these cells are deficient in producing nutrients required for their growth. By supplying the gene for the deficient nutrient in the 'bait' or 'hunter' plasmid, cells containing the plasmid are able to survive on the minimal media, whereas untransfected cells cannot (Figure 5). Selection in this way occurs in two 21 rounds: first on one minimal media plate, to select for the 'bait' plasmid, and then on another minimal media plate, to select for the 'hunter'4. Once inside the cell, if binding occurs between the hunter and the bait, transcriptional activity will be restored and will produce normal Gal4 activity. The reporter gene most commonly used in the Gal4 system is LacZ, an E. coli gene whose transcription causes cells to turn blue4. In this yeast system, the LacZ gene is inserted in the yeast DNA immediately after the Gal4 promoter, so that if binding occurs, LacZ is produced. Therefore, detecting interactions between bait and hunter simply requires identifying blue versus non-blue. 22