* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Cells (Ch3)
Cell encapsulation wikipedia , lookup
Cellular differentiation wikipedia , lookup
Magnesium transporter wikipedia , lookup
Cell growth wikipedia , lookup
SNARE (protein) wikipedia , lookup
Extracellular matrix wikipedia , lookup
Cell nucleus wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Cytokinesis wikipedia , lookup
Signal transduction wikipedia , lookup
Cell membrane wikipedia , lookup
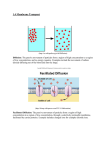
CELLS Chapter 3 Cell Theory • Cell: structural and functional unit of life • Organismal functions depend on individual and collective cell functions • Biochemical activities of cells dictated by their shapes or forms, and specific subcellular structures • Continuity of life has cellular basis Cell Diversity • Over 200 different types of human cells • Types differ in size, shape, subcellular components, and functions Plasma Membrane • Bimolecular layer of lipids and proteins in a constantly changing fluid mosaic • Plays a dynamic role in cellular activity • Separates intracellular fluid (ICF) from extracellular fluid (ECF) – Interstitial fluid (IF) = ECF that surrounds cells Membrane Lipids • 75% phospholipids (lipid bilayer) – Phosphate heads: polar and hydrophilic – Fatty acid tails: nonpolar and hydrophobic • 5% glycolipids – Lipids with polar sugar groups on outer membrane surface • 20% cholesterol – Increases membrane stability and fluidity • Lipid Rafts – ~ 20% of the outer membrane surface – Contain phospholipids, sphingolipids, and cholesterol – May function as stable platforms for cell-signaling molecules Membrane Proteins • Integral proteins – Firmly inserted into the membrane (most are transmembrane) – Functions: Transport proteins (channels and carriers), enzymes, or receptors • Peripheral Proteins – Loosely attached to integral proteins – Include filaments on intracellular surface and glycoproteins on extracellular surface – Functions: Enzymes, motor proteins, cell-to-cell links, provide support on intracellular surface, and form part of glycocalyx = protein = lipid Extracellular fluid (watery environment) Polar head of phospholipid molecule Cholesterol Glycolipid Glycoprotein Carbohydrate of glycocalyx Outwardfacing layer of phospholipids Integral proteins Filament of cytoskeleton Peripheral Bimolecular Inward-facing proteins lipid layer layer of containing phospholipids Nonpolar proteins tail of phospholipid Cytoplasm molecule (watery environment) Figure 3.3 Cell Junctions • Some cells "free” • Some bound into communities – Three ways cells are bound: • Tight junctions • Desmosomes • Gap junctions Tight Junctions • Adjacent integral proteins fuse form impermeable junction encircling cell – Prevent fluids and most molecules from moving between cells Desmosomes • Anchor cells together at plaques (thickenings on plasma membrane) – Linker proteins between cells connect plaques – Keratin filaments (part of the cytoskeleton) extend through cytosol to opposite plaque giving stability to cell – Reduces possibility of tearing Gap Junctions • Transmembrane proteins form pores (connexons) that allow small molecules to pass from cell to cell – For spread of ions, simple sugars, and other small molecules between cardiac or smooth muscle cells Cell Cycle • Defines changes from formation of cell until it reproduces • Includes: – Interphase – Cell division (mitotic phase) Interphase • Period from cell formation to cell division • Nuclear material called chromatin • Three subphases: – G1 (gap 1)—vigorous growth and metabolism • Cells that permanently cease dividing said to be in G0 phase – S (synthetic)—DNA replication occurs – G2 (gap 2)—preparation for division © 2013 Pearson Education, Inc. Interphase Centrosomes (each has 2 centrioles) Plasma membrane Nucleolus Chromatin Nuclear envelope © 2013 Pearson Education, Inc. DNA Replication Cell Division • Meiosis - cell division producing gametes • Mitotic cell division - produces clones – Essential for body growth and tissue repair – Occurs continuously in some cells • Skin; intestinal lining – None in most mature cells of nervous tissue, skeletal muscle, and cardiac muscle • Repairs with fibrous tissue Events Of Cell Division • Mitosis—division of nucleus – Four stages • • • • Prophase Metaphase Anaphase Telophase – Cytokinesis—division of cytoplasm-by cleavage furrow Prophase Metaphase Anaphase Telophase and Cytokinesis Telophase Cytokinesis Nuclear envelope forming Nucleolus forming Contractile ring at cleavage furrow Control of Cell Division • "Go" signals: – Critical volume of cell when area of membrane inadequate for exchange – Chemicals (e.g., growth factors, hormones) – Availability of space–contact inhibition • To replicate DNA and enter mitosis requires – Cyclins–regulatory proteins • Accumulate during interphase – Cdks (Cyclin-dependent kinases)–bind to cyclins activated • Enzyme cascades prepare cell for division – Cyclins destroyed after mitotic cell division Control of Cell Division • Checkpoints – G1 checkpoint (restriction point) most important • If doesn't pass G0–no further division – Late in G2 MPF (M-phase promoting factor) required to enter M phase • "Other Controls" signals – Repressor genes inhibit cell division • E.g., P53 gene Protein Synthesis Overview Two Components • Transcription • Translation RNA is heavily involved Roles of the Three Main Types of RNA • Messenger RNA (mRNA) – Carries instructions for building a polypeptide, from gene in DNA to ribosomes in cytoplasm • Ribosomal RNA (rRNA) – A structural component of ribosomes that, along with tRNA, helps translate message from mRNA • Transfer RNAs (tRNAs) – Bind to amino acids and pair with bases of codons of mRNA at ribosome to begin process of protein synthesis Transcription • Transfers DNA gene base sequence to a complementary base sequence of an mRNA • Transcription factor – Loosens histones from DNA in area to be transcribed – Binds to promoter, a DNA sequence specifying start site of gene to be transcribed – Mediates the binding of RNA polymerase to promoter Transcription • RNA polymerase – Enzyme that oversees synthesis of mRNA – Unwinds DNA template – Adds complementary RNA nucleotides on DNA template and joins them together – Stops when it reaches termination signal – mRNA pulls off the DNA template, is further processed by enzymes, and enters cytosol RNA polymerase Transcription Coding strand DNA Promoter region Template strand Termination signal 1 Initiation: With the help of transcription factors, RNA polymerase binds to the promoter, pries apart the two DNA strands, and initiates mRNA synthesis at the start point on the template strand. mRNA Template strand Coding strand of DNA 2 Elongation: As the RNA polymerase moves along the template Rewinding of DNA strand, elongating the mRNA transcript one base at a time, it unwinds the DNA double helix before it and rewinds the double helix behind it. mRNA transcript RNA nucleotides Direction of transcription mRNA DNA-RNA hybrid region Template strand RNA polymerase 3 Termination: mRNA synthesis ends when the termination signal is reached. RNA polymerase and the completed mRNA transcript are released. Unwinding of DNA The DNA-RNA hybrid: At any given moment, 16–18 base pairs of DNA are unwound and the most recently made RNA is still bound to DNA. This small region is called the DNA-RNA hybrid. Completed mRNA transcript RNA polymerase Figure 3.35 Translation • Converts base sequence of nucleic acids into the amino acid sequence of proteins • Involves mRNAs, tRNAs, and rRNAs • Each three-base sequence on DNA is represented by a codon – Codon—complementary three-base sequence on mRNA – Each codon corresponds to a specific amino acid What do you notice about this? How many stop codons are there? Start codons? Translation • mRNA attaches to a small ribosomal subunit that moves along the mRNA to the start codon • Large ribosomal unit attaches, forming a functional ribosome • Anticodon of a tRNA binds to its complementary codon and adds its amino acid to the forming protein chain • New amino acids are added by other tRNAs as ribosome moves along rRNA, until stop codon is reached Translation Role of Rough ER in Protein Synthesis • mRNA–ribosome complex is directed to rough ER by a signal-recognition particle (SRP) • Forming protein enters the ER • Sugar groups may be added to the protein, and its shape may be altered • Protein is enclosed in a vesicle for transport to Golgi apparatus 1 The mRNA-ribosome complex is directed to the rough ER by the SRP. There the SRP binds to a receptor site. ER signal sequence 2 Once attached to the ER, the SRP is released and the growing polypeptide snakes through the ER membrane pore into the cisterna. 3 The signal sequence is clipped off by an enzyme. As protein synthesis continues, sugar groups may be added to the protein. Ribosome mRNA Signal Signal recognition sequence particle Receptor site removed (SRP) Growing polypeptide 4 In this example, the completed protein is released from the ribosome and folds into its 3-D conformation, a process aided by molecular chaperones. Sugar group 5 The protein is enclosed within a protein (coatomer)-coated transport vesicle. The transport vesicles make their way to the Golgi apparatus, where further processing of the proteins occurs (see Figure 3.19). Released protein Rough ER cisterna Cytoplasm Transport vesicle pinching off Coatomer-coated transport vesicle Figure 3.39 Membrane Transport • Plasma membranes are selectively permeable • Passive Processes – No cellular energy (ATP) required – Substance moves down its concentration gradient • Active Processes – Energy (ATP) required – Occurs only in living cell membranes Passive Processes • Simple diffusion – Nonpolar lipid-soluble (hydrophobic) substances diffuse directly through the phospholipid bilayer • Facilitated diffusion – Carrier or channel mediated • Osmosis Passive Processes: Facilitated Diffusion • Some lipophobic molecules (e.g., glucose, amino acids, and ions) use carrier proteins or channel proteins, both of which: – Exhibit specificity (selectivity) – Are saturable; rate is determined by number of carriers or channels – Can be regulated in terms of activity and quantity Facilitated Diffusion Using Carrier Proteins • Transmembrane integral proteins transport specific polar molecules (e.g., sugars and amino acids) • Binding of substrate causes shape change in carrier Facilitated Diffusion Using Channel Proteins • Aqueous channels formed by transmembrane proteins selectively transport ions or water • Two types: – Leakage channels • Always open – Gated channels • Controlled by chemical or electrical signals Passive Processes: Osmosis • Movement of solvent (water) across a selectively permeable membrane • Water diffuses through plasma membranes: – Through the lipid bilayer – Through water channels called aquaporins (AQPs) Passive Processes: Osmosis • Water concentration is determined by solute concentration • Osmolarity: total concentration of solute particles • When solutions of different osmolarity are separated by a membrane, osmosis occurs until equilibrium is reached • In Cells: When osmosis occurs, water enters or leaves a cell – Change in cell volume disrupts cell function Tonicity • Tonicity: The ability of a solution to cause a cell to shrink or swell • Isotonic: A solution with the same solute concentration as that of the cytosol • Hypertonic: A solution having greater solute concentration than that of the cytosol • Hypotonic: A solution having lesser solute concentration than that of the cytosol Membrane Transport: Active Processes • Two types of active processes: – Active transport – Vesicular transport • Both use ATP to move solutes across a living plasma membrane Active Transport • Requires carrier proteins (solute pumps) • Moves solutes against a concentration gradient • Types of active transport: – Primary active transport – Secondary active transport Primary Active Transport • Hydrolysis of ATP (energy!) causes shape change in transport protein • bound solutes (ions) are “pumped” across the membrane • Sodium-potassium pump (Na+-K+ ATPase) – Located in all plasma membranes – Involved in primary (and secondary) active transport of nutrients and ions – Maintains electrochemical gradients essential for functions of muscle and nerve tissues Secondary Active Transport • Depends on an ion gradient created by primary active transport • Energy stored in ionic gradients is used indirectly to drive transport of other solutes • Cotransport—always transports more than one substance at a time – Symport system: Two substances transported in same direction – Antiport system: Two substances transported in opposite directions Extracellular fluid Na+-K+ pump Cytoplasm 1 The ATP-driven Na+-K+ pump stores energy by creating a steep concentration gradient for Na+ entry into the cell. Figure 3.11 step 1 Extracellular fluid Glucose Na+-K+ pump Na+-glucose symport transporter loading glucose from ECF Na+-glucose symport transporter releasing glucose into the cytoplasm Cytoplasm 1 The ATP-driven Na+-K+ pump 2 As Na+ diffuses back across the stores energy by creating a steep concentration gradient for Na+ entry into the cell. membrane through a membrane cotransporter protein, it drives glucose against its concentration gradient into the cell. (ECF = extracellular fluid) Figure 3.11 step 2 Vesicular Transport • Transport of large particles, macromolecules, and fluids across plasma membranes • Requires cellular energy (e.g., ATP) • Functions: – Exocytosis—transport out of cell – Endocytosis—transport into cell – Transcytosis—transport into, across, and then out of cell – Substance (vesicular) trafficking—transport from one area or organelle in cell to another Endocytosis and Transcytosis • Involve formation of protein-coated (typically clathrin) vesicles • Often receptor mediated, therefore very selective http://www.biologycorner.com/resources/endocytosis.gif 1 Coated pit ingests substance. Extracellular fluid Protein coat (typically clathrin) 2 Proteincoated vesicle detaches. Plasma membrane Cytoplasm 3 Coat proteins detach and are recycled to plasma membrane. Transport vesicle Endosome Uncoated endocytic vesicle 4 Uncoated vesicle fuses with a sorting vesicle called an endosome. Lysosome 5 Transport vesicle containing membrane components moves to the plasma membrane for recycling. 6 Fused vesicle may (a) fuse (a) with lysosome for digestion of its contents, or (b) deliver its contents to the plasma membrane on the opposite side of the cell (transcytosis). (b) Figure 3.12 Endocytosis • Phagocytosis—pseudopods engulf solids and bring them into cell’s interior – Macrophages and some white blood cells Endocytosis • Fluid-phase endocytosis (pinocytosis)— plasma membrane infolds, bringing extracellular fluid and solutes into interior of the cell – Nutrient absorption in the small intestine Endocytosis • Receptor-mediated endocytosis— highly selective – Uptake of enzymes low-density lipoproteins, iron, and insulin Vesicle Receptor recycled to plasma membrane (c) Receptor-mediated endocytosis Extracellular substances bind to specific receptor proteins in regions of coated pits, enabling the cell to ingest and concentrate specific substances (ligands) in protein-coated vesicles. Ligands may simply be released inside the cell, or combined with a lysosome to digest contents. Receptors are recycled to the plasma membrane in vesicles. Figure 3.13c Exocytosis • Examples: – Hormone secretion – Neurotransmitter release – Mucus secretion – Ejection of wastes Plasma membrane The process Extracellular of exocytosis SNARE (t-SNARE) fluid Secretory vesicle Vesicle SNARE (v-SNARE) Molecule to be secreted Cytoplasm 1 The membrane- bound vesicle migrates to the plasma membrane. 2 There, proteins at the vesicle Fused surface (v-SNAREs) v- and bind with t-SNAREs t-SNAREs (plasma membrane proteins). Fusion pore formed 3 The vesicle and plasma membrane fuse and a pore opens up. 4 Vesicle contents are released to the cell exterior. Figure 3.14a Membrane Potential • Separation of oppositely charged particles (ions) across a membrane creates a membrane potential (potential energy measured as voltage) • Resting membrane potential (RMP): Voltage measured in resting state in all cells – Ranges from –50 to –100 mV in different cells – Results from diffusion and active transport of ions (mainly K+) 1 K+ diffuse down their steep Extracellular fluid concentration gradient (out of the cell) via leakage channels. Loss of K+ results in a negative charge on the inner plasma membrane face. 2 K+ also move into the cell because they are attracted to the negative charge established on the inner plasma membrane face. 3 A negative membrane potential Potassium leakage channels Cytoplasm (–90 mV) is established when the movement of K+ out of the cell equals K+ movement into the cell. At this point, the concentration gradient promoting K+ exit exactly opposes the electrical gradient for K+ entry. Protein anion (unable to follow K+ through the membrane) Figure 3.15