* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Microbial metabolism
Nicotinamide adenine dinucleotide wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Metalloprotein wikipedia , lookup
Paracrine signalling wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Signal transduction wikipedia , lookup
Biosynthesis wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Photosynthesis wikipedia , lookup
Citric acid cycle wikipedia , lookup
Biochemical cascade wikipedia , lookup
Phosphorylation wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Biochemistry wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Electron transport chain wikipedia , lookup
Microbial metabolism wikipedia , lookup
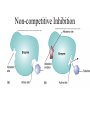
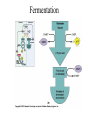
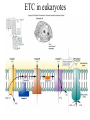
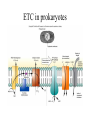
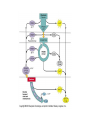
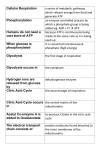
Energy is the capacity to do work • Potential energy: stored energy • Kinetic energy: energy of motion Metabolism • Metabolism is the sum total of ALL chemical reactions within a cell – Catabolic – Anabolic / Biosynthetic A metabolic pathway is a series of sequential, interconnected, enzyme-catalyzed biochemical reactions by which a living cell converts a starting product (substrate) into an end product. Metabolism • Catabolism: “destructive metabolism” – Breakdown of substrate: end product is less complex than the starting product – Catabolic pathways generate energy and precursor metabolites – Catabolic pathways are linked to either respiration or fermentation • Anabolism = biosynthesis: “constructive metabolism” – Synthesis of new biomolecules: end product is more complex than the starting product – Anabolic pathways use energy and precursor metabolites to synthesize subunits of macromolecules Adenosine triphosphate: ATP • The main energy currency of the cell • Ribose + adenine + 3 phosphate groups • Phosphate bonds = high energy bonds: energy is used to form the bonds and released when the bonds are broken • Addition of a phosphate group to ADP to generate ATP is accomplished during metabolism via either substrate level phosphorylation or oxidative phosphorylation And microorganisms too! ATP is made in catabolic reactions and used in anabolic reactions Ways cells make ATP • Substrate level phosphorylation – used by all organisms; for fermenters, this is the only option • Oxidative phosphorylation – aerobic and anaerobic respiration • Photophosphorylation – photosynthesizers only Products of Anabolic Metabolism (Biosynthesis) • Fatty acids • Glycerol – Fatty acids + glycerol = lipids (where do you find lipid in a bacterial cell?) • Amino acids – Building blocks for proteins • Nucleotides – Building blocks for DNA, RNA The critical components of metabolic pathways are: • • • • • Enzymes Adenosine triphosphate (ATP) Chemical energy source Terminal electron acceptor Electron carriers Oxidation-Reduction Rections (redox reactions) • Same principal in biological systems as inorganic systems: – Substance that gives up (loses) electron(s) = oxidized – Substance that gains electron(s) = reduced • When electrons move, protons (H+) follow Oxidation/reduction reactions Biological Oxidation Enzymes • All enzymes are proteins • Active site = binding site: highly substrate-specific • Binding of the substrate induces a conformational change in the enzyme – This forms the temporary enzyme-substrate complex – Formation of this complex changes the position of bonds on the substrate, destabilizing them and thereby lowering the activation energy of the reaction • When the changed substrate is released, the enzyme returns to the native conformation and is ready to bind another substrate – the enzyme is chemically unchanged by the reaction. Enzymes • Active site: site that binds the substrate • Allosteric sites: binding site at a location separate from the active site – binding to the allosteric site can induce a conformational change in the active site • Allosteric regulation: binding of a molecule to the allosteric site to either allow or block binding of the substrate – This allows the cell to regulate pathways based on the quantity of the end product available, so they only make what they need • Coenzymes: organic cofactors that assist in the transfer of molecules or electrons • Environmental factors that influence enzymatic function/activity: – temperature, pH, salt concentration Factors that influence an enzyme: Temperature • What happens as temperature increases? • What is the optimum temperature? Factors that influence an enzyme: pH • What pH do most enzymes function optimally? Enzymes bind substrate and generate a product Some enzymes require a cofactor to bind substrate Coenzymes carry electrons Enzyme inhibitors • Inhibit the binding of the substrate to the active site – Competitive inhibition – Non-Competitive Inhibition Competitive Inhibition Non-competitive Inhibition Electron carriers • Special types of coenzymes that transfer electrons (and protons) between molecules • NAD+/NADH: nicotinamide adenine dinucleotide • NADP+/NADPH: nicotinamide adenine dinucleotide phosphate • FAD/FADH2: flavin adenine dinucleotide Electron Transport Chain & Proton Motive Force • The ETC is an association of electron carriers located within a cell membrane. • As electrons pass between electron carriers in the ETC they move from higher to lower affinity carriers, releasing energy in the process. • Movement of electrons is paired with the movement of protons across the membrane to generate an electrical gradient • This electrical gradient = the proton motive force. • It consists of a positive charge at the outside surface of the membrane and a negative charge at the inner surface • The electrical potential energy of the proton motive force can be converted to chemical energy = production of ATP The Electron Transport Chain and the Proton Motive Force: Prokaryotic vs eukaryotic cells • Prokaryotic cells: – Electron transport chain is located in the cytoplasmic membrane – Electrical gradient of the proton motive force is across the cell membrane (inside vs outside cell) – Proton motive force can be used to generate ATP, or to directly power flagella or transport molecules against the concentration gradient (active transport) • These latter 2 processes can also be fueled by ATP generated via the proton motive force • Eukaryotic cells: – ETC is located in the inner mitochondrial membrane – Electrical gradient of proton motive force is between the matrix and the region between the inner and outer membranes – Only function of the proton motive force is generation of ATP Central metabolic pathways: catabolic pathways • Glycolysis: primary pathway for most organisms • Substrate = glucose • End product = pyruvate • The other pathways are the pentose phosphate pathway and the tricarboxylic acid pathway – these are linked by a “transition step” Types of Bacterial Metabolism • Fermentation • Respiration – Aerobic Respiration – Anaerobic Respiration • Photosynthesis Comparison of three types of metabolism make a new slide to replace this = a table summarizing what I want them to know = name of the process, pathways used, terminal electron accentor and “energy yield” – substrate level + phosphorylation and oxidative phosphorylation – need a slide summarizing the difference between these two – also a note on this slide saying I just want them to know that for both SLP and OP the energy yielod is greatest for aerobic respiration, lowest for fermentation and intermediate for anaerobic respiration; and also that in fermentation there is ONLY substrate level phosphorylation, not oxidative phosphorylation Cellular Respiration • The series of steps by which cells produce energy through oxidation of organic substances • At a cellular level, this can be linked to catabolism – electrons extracted in the metabolism of glucose (or other substrates) are transferred to the electron transport chain to generate ATP via oxidative phosphorylation Respiration • Aerobic respiration: – Uses ETC – Oxygen is the terminal electron acceptor – ATP is generated by both substrate level and oxidative phosphorylation – Higher energy yield than anaerobic respiration or fermentation • Anaerobic respiration: – Uses ETC – Terminal electron acceptor = various molecules other than O2, ie nitrate, nitrite, sulfate – ATP is generated by both substrate level and oxidative phosphorylation Aerobic Respiration • The COMPLETE breakdown of glucose to CO2 and H2O with an inorganic compound serving as the final electron acceptor Remember the pathways in aerobic respiration are… • Glycolysis – Some use Pentose Phosphate Pathway instead • TCA cycle • Electron transport chain Fermentation • The incomplete breakdown of glucose with an organic compound serving as the final electron acceptor • Generates less energy than aerobic or anerobic respiration • Only central metabolic pathway operating is glycolysis • Question: if fermentation is so inefficient, why do cells use this pathway? Fermentation products can vary Fermentation Saccharomyces produces ethanol Lactic Acid Bacteria Precursor metabolites • There are many important precursor metabolites formed during the central metabolic pathways which are used in biosynthetic (anabolic) pathways. – Fructose 6-phosphate • Generated during glycolysis • Used in the synthesis of peptidoglycan – Glucose 6-phosphate • Generated during glycolysis • Used in the synthesis of lipopolysaccharide Carbon fixation • Organism that are able to use CO2 as a carbon source = • Pathways utilized include Calvin Cycle and reverse TCA cycle • Some bacteria are able to break down CO2 to yield carbon Chemoorganotrophs depend on Photosynthetic organisms What is made as a result of the TCA cycle? • ATP • Reducing power • Precursor metabolites made from alphaketoglutarate and oxaloacetate Electron Transport Chain • Found in the cytoplasmic membrane • Contains electron carriers – – – – Flavoproteins Iron-sulfur proteins Quinones Cytochromes Model for energy release in ETC ETC in eukaryotes ETC in prokaryotes ATP yield from aerobic respiration Remember we are focusing on catabolic reactions • Generate ATP for later use by cell • Generate precursors for other pathways • Need to re-oxidize coenzymes for continual use