* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download A Tour of the Cell Chapter 6: 1. Studying Cells 2. Intracellular Structures
Survey
Document related concepts
Cytoplasmic streaming wikipedia , lookup
Cell growth wikipedia , lookup
Tissue engineering wikipedia , lookup
Cellular differentiation wikipedia , lookup
Signal transduction wikipedia , lookup
Cell encapsulation wikipedia , lookup
Cell culture wikipedia , lookup
Cell membrane wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Extracellular matrix wikipedia , lookup
Cell nucleus wikipedia , lookup
Cytokinesis wikipedia , lookup
Transcript
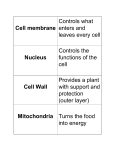
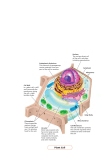
Chapter 6: A Tour of the Cell 1. Studying Cells 2. Intracellular Structures 3. The Cytoskeleton 4. Extracellular Structures 1. Studying Cells 1m Human height Length of some nerve and muscle cells 0.1 m Chicken egg 1 cm Frog egg 1 mm 100 µm RESOLUTION • minimum distance across which 2 points can be resolved or seen distinctly (limited by wavelength) CONTRAST • degree to which objects differ from background Most plant and animal cells 10 µm Nucleus Most bacteria 1 µm 100 nm 10 nm Mitochondrion Smallest bacteria Viruses Ribosomes Proteins Lipids 1 nm Small molecules 0.1 nm Atoms Electron microscope • factor by which the image produced is larger than the actual object (e.g. “100X”) 10 m Light microscope MAGNIFICATION Limits of Resolution Unaided eye Concepts of Microscopy Light Microscopy a typical compound microscope such as used in your lab Bright Field Microscopy TECHNIQUE RESULTS (a) Brightfield (unstained specimen) Standard form of Light Microscopy, poor contrast 50 µm (b) Brightfield (stained specimen) Staining increases contrast, though the staining process usually kills the specimen Phase Contrast Microscopy TECHNIQUE (c) Phase-contrast Enhances misalignment of light waves to create contrast Reveals internal detail without staining, useful for viewing live specimens (d) Differential-interferencecontrast (Nomarski) A variation of phase-contrast microscopy involving a more complex combination of filters and prisms. RESULTS Fluorescence Microscopy TECHNIQUE RESULTS (e) Fluorescence Fluorescent dyes or antibodies with a fluorescent tag stick to specific targets which then fluoresce under UV light. 50 µm Only the objects or structures that fluoresce are visible. • objects that bind the fluorescent stain or antibody • objects that are naturally fluorescent Antibodies Proteins made by B cells that bind to a unique antigen: • the variable (V) region recognizes specific antigen • the constant (C) region is the same for all Ab’s in a given class: IgM IgD IgA IgG IgE (Ig = “immunoglobulin”) Confocal Fluorescence Microscopy Only light from a given depth or plane is transmitted, “out of focus” light is excluded TECHNIQUE (a) Scanning electron microscopy (SEM) RESULTS Cilia 1 µm Electromagnetic lenses focus electron beam onto heavy metalstained specimen. view of whole specimen, reveals surface features (b) Transmission electron microscopy (TEM) specimen cut in thin sections, higher resolution Electron Microscopy Longitudinal section of cilium Cross section of cilium 1 µm • electron beams have very short wavelengths • allows far greater resolution than with light microscopy TECHNIQUE Fractionation by Centrifugation In addition to microscopic examination, cells and their structures are also studied biochemically: • in order to study a cellular compartment biochemically, it must be separated from the rest of the cell • this is accomplished through successive centrifugation steps at increasing speeds Homogenization Tissue cells 1,000 g (1,000 times the force of gravity) 10 min Homogenate Differential centrifugation Supernatant poured into next tube 20,000 g 20 min 80,000 g 60 min Pellet rich in nuclei and cellular debris 150,000 g 3 hr Pellet rich in mitochondria (and chloroplasts if cells are from a plant) Pellet rich in “microsomes” (pieces of plasma membranes and cells’ internal membranes) Pellet rich in ribosomes Surface area increases while total volume remains constant Why are cells the size they are? 5 1 1 Total surface area [Sum of the surface areas (height width) of all boxes sides number of boxes] Total volume [height width length number of boxes] Surface-to-volume (S-to-V) ratio [surface area ÷ volume] 6 150 750 1 125 125 6 1.2 6 Why aren’t cells bigger? 1) Cell size is limited by the rate of diffusion: • if cells get too large, it takes too much time for nutrients, wastes, etc, to disperse in the cell 2) And also by the surface to volume ratio (S/V): surface area of sphere = 4pr2 volume of sphere = (4/3)pr3 *Surface Area increases by square of radius *Volume increases by cube of radius ***The larger the cell, the smaller the S/V ratio*** 2. Intracellular Structures Cells come in 2 basic types: 1. Prokaryotic cells (“before” nucleus) • lack a nucleus & other organelles • small, unicellular diameter ~1-10 mm • organisms in the following domains: Bacteria Archaea 2. Eukaryotic cells (“true” nucleus) • have a nucleus, subcellular organelles • unicellular or multicellular • “large” (diameter ~10 mm – 1 mm) • Eukarya: Protists, Fungi , Plants & Animals Prokaryotic Cells Fimbriae Nucleoid Ribosomes Plasma membrane Bacterial chromosome Cell wall Capsule 0.5 µm (a) A typical rodshaped bacterium Flagella (b) A thin section through the bacterium Bacillus coagulans (TEM) Have intracellular organization despite no organelles. * Nuclear envelope not in plant cells ENDOPLASMIC RETICULUM (ER) Nucleolus * * Rough ER NUCLEUS Smooth ER Flagellum Chromatin Centrosome Plasma membrane CYTOSKELETON: Microfilaments Intermediate filaments Microtubules Ribosomes * Microvilli Golgi apparatus Peroxisome Mitochondrion * Animal Cell Lysosome The Nucleus Where genetic material (DNA) is stored, gene expression begins. Nucleus 1 µm Nucleolus Chromatin Nucleolus Nuclear envelope: Inner membrane Outer membrane where ribosomal subunits are assembled from rRNA & proteins Nuclear pore Pore complex Surface of nuclear envelope Rough ER Ribosome 1 µm 0.25 µm complex of DNA & histone proteins Close-up of nuclear envelope Pore complexes (TEM) Chromatin Nuclear lamina (TEM) Ribosomes Carry out protein synthesis by the process of translation. Cytosol Endoplasmic reticulum (ER) Free ribosomes Bound ribosomes Large subunit 0.5 µm TEM showing ER and ribosomes Small subunit Diagram of a ribosome Smooth ER Rough ER Nuclear envelope Endoplasmic Reticulum Rough ER (RER) ER lumen Cisternae Ribosomes Transport vesicle Smooth ER Transitional ER Rough ER • ribosomes on cytoplasmic face of ER membrane synthesize proteins across ER membrane into lumen of ER 200 nm • beginning of the secretory pathway Smooth ER (SER) • has membrane-associated enzymes that catalyze new lipid synthesis (also found in RER), neutralizing toxins • storage of calcium ions The Golgi apparatus cis face (“receiving” side of Golgi apparatus) 0.1 µm Cisternae trans face (“shipping” side of Golgi apparatus) TEM of Golgi apparatus • proteins destined to leave ER are transported to the Golgi where they are modified, sorted and sent to various destinations. • polysaccharides are produced in the Golgi apparatus as well Lysosomes Nucleus 1 µm Vesicle containing two damaged organelles 1 µm Mitochondrion fragment Peroxisome fragment Lysosome Lysosome Digestive enzymes Lysosome Plasma membrane Peroxisome Digestion Food vacuole Vesicle (a) Phagocytosis Mitochondrion Digestion (b) Autophagy • acidic compartments full of enzymes for the breakdown or digestion of foreign or waste material The Endomembrane System Nucleus Rough ER Smooth ER cis Golgi aka the “Secretory Pathway” trans Golgi Plasma membrane Mitochondria Have ribosomes that resemble those of prokaryotes Intermembrane space Outer membrane Free ribosomes in the mitochondrial matrix Inner membrane Cristae Matrix 0.1 µm ATP production via cellular respiration • convert energy from glucose, fatty acids, etc, to energy in ATP NUCLEUS Nuclear envelope Nucleolus Chromatin Rough endoplasmic reticulum *not in animal cells Smooth endoplasmic reticulum Ribosomes * Central vacuole Golgi apparatus Microfilaments Intermediate filaments CYTOSKELETON Microtubules Mitochondrion Peroxisome Chloroplast Plasma membrane * Cell wall Wall of adjacent cell * * Plasmodesmata Plant Cell Central Vacuole Plant organelle that stores water and various ions Central vacuole Source of “turgor pressure” that maintains rigidity of plant cells Cytosol Nucleus • swells when water is plentiful due to osmosis Central vacuole Cell wall Chloroplast 5 µm • cell wall provides support, prevents lysis Chloroplasts Site of photosynthesis in plant cells. Chloroplast Like mitochondria, have ribosomes and other components that resemble those of prokaryotes Stroma Inner and outer membranes Granum Intermembrane space • production of glucose from CO2 and H2O using sunlight • the basis of essentially all ecosystems Peroxisomes Chloroplast Peroxisome Mitochondrion Contain enzymes that oxidize (i.e., remove H) various organic molecules thus forming H2O2 from O2 • H2O2 is then converted to O2 and H2O 1 µm Involved in the detoxification of toxic substances, breakdown of fatty acids 3. The Cytoskeleton The Cytoskeleton A complex, highly dynamic intracellular network of protein filaments largely responsible for: • cell movement, motility • cell shape, rigidity • movement of vesicles (and organelles) Microtubule • localization of organelles • dynamics of cell division 0.25 µm Microfilaments 3 Basic Cytoskeletal Filaments Actin filaments/microfilaments (MF) intermediate filaments (IF) microtubules (MT) MF MT IF Microfilaments 10 µm Actin subunit 7 nm Intermediate Filaments 5 µm Keratin proteins Fibrous subunit (keratins coiled together) 8–12 nm 10 µm Microtubules Column of tubulin dimers 25 nm Tubulin dimer ATP Vesicle Transport Vesicle Receptor for motor protein Motor protein (ATP powered) Microtubule of cytoskeleton (a) Microtubule (b) Vesicles 0.25 µm • a variety of motor proteins are involved in binding and transporting vesicles along cytoskeletal fibers to their destination Centrosome Centrosomes Microtubule Centrioles • centrosomes contain a pair of centrioles (animal cells only) 0.25 µm • these structures are involved in the formation of the mitotic spindle or “spindle fibers” that play such an important role in cell division Longitudinal section of one centriole Microtubules Cross section of the other centriole Flagella & Cilia FLAGELLA are involved in cell motility, are very long, and cells have relatively few (1 or several) Direction of swimming (a) Motion of flagella 5 µm Direction of organism’s movement Power stroke Recovery stroke (b) Motion of cilia 15 µm CILIA are involved in motility, moving material across the cell surface, and are present on the cell surface in high numbers 4. Extracellular Structures Plant Cell Walls Cell walls Interior of cell Secondary cell wall Primary cell wall Interior of cell 0.5 µm Plasmodesmata Middle lamella Plasma membranes Plant cell walls contain fibers of cellulose and other polysaccharides as well as proteins 1 µm Central vacuole Cytosol Plasma membrane Plant cell walls • one or more layers of secondary cell wall may be produced in some plant cells Plasmodesmata Intercellular Junctions Tight junction Tight junctions prevent fluid from moving across a layer of cells TIGHT JUNCTIONS are impenetrable seals connecting adjacent cells that prevent fluid and other materials from passing between the cells 0.5 µm Tight junction Intermediate filaments DESMOSOMES are strong connections between cells that create a very strong sheet of cells GAP JUNCTIONS (animal) & PLASMODESMATA (plant) provide channels through which ions & other small molecules can pass from cell to cell Desmosome Desmosome Gap junctions Space between cells Plasma membranes of adjacent cells Extracellular matrix 1 µm Gap junction 0.1 µm The Extracellular Matrix A meshwork of protein fibers and polysaccharides that retain fluid and produce gel-like matrix that holds cells together in tissues. Polysaccharide molecule Collagen Proteoglycan complex EXTRACELLULAR FLUID Carbohydrates Core protein Fibronectin Integrins Plasma membrane Proteoglycan molecule Microfilaments CYTOPLASM Proteoglycan complex Key Terms for Chapter 6 • magnification, resolution, contrast • bright field, phase contrast, fluorescent, confocal, transmission & scanning electron microscopy • prokaryotic vs eukaryotic • cell wall, capsule, flagella, nucleoid, cytoplasm • nucleus, nucleolus, endoplasmic reticulum, ribosome • Golgi apparatus, lysosome, peroxisome, vesicle • endomembrane system, central vacuole • mitochondria, chloroplasts • cytoskeleton, cilia, flagella, centrosome, centriole • microtubules, microfilaments, intermediate filaments • motor proteins • tight junctions, gap junctions, desmosomes, plasmodesmata • extracellular matrix, proteoglycan Relevant Chapter Questions 1-9