* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download 1/25
Skewed X-inactivation wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Ridge (biology) wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Neocentromere wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Public health genomics wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Gene desert wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Gene therapy wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Oncogenomics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Minimal genome wikipedia , lookup
Gene nomenclature wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic engineering wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genome editing wikipedia , lookup
Helitron (biology) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene expression programming wikipedia , lookup
Epigenetics of human development wikipedia , lookup
X-inactivation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene expression profiling wikipedia , lookup
Point mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome (book) wikipedia , lookup
Designer baby wikipedia , lookup
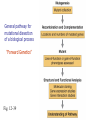
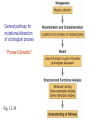
Mutagenesis and Genetic Screens General pathway for mutational dissection of a biological process “Forward Genetics” Fig. 12-39 From phenotype to gene • Once an interesting mutant is found and chromosome characterized, we want to find the gene in which the mutant occurred • Positional cloning – First use genetic mapping – Then use chromosome walking contig candidate genes mutation Candidate-gene approach • If the mutated gene is localized to a sequenced region of the chromosome, then look for genes that could be involved in the process under study • Last step: confirm gene identification – Rescue of phenotype – Mutations in same gene in different alleles Insertional mutagenesis • Alternative to chromosome walking – To reduce time and effort required to identify mutant gene • Insert piece of DNA that disrupts genes – Inserts randomly in chromosomes • Make collection of individuals – Each with insertion in different place • Screen collection for phenotypes • Use inserted DNA to identify mutated gene General pathway for mutational dissection of a biological process “Forward Genetics” Fig. 12-39 Isolated 148 temperature-sensitive cell division cycle (cdc) mutants out of 1500 total ts mutants Complementation analysis: 32 complementation groups (genes/cistrons) Mapped 14 genes (no apparent clustering) Phenotypic characterizations: •Variety of cell cycle points disrupted (can dissect stages) •Mutations of single gene blocked in very similar point (same morphology indicates action at a single point)) •Temperature shift immediately blocks some mutants; block delayed 1-3 cell cycles for others (suggests temperature effects are on protein function versus protein synthesis) •Mutants block cell cycle in haploid and diploid cells 2 cdc 2 marker 2 cdc & marker 2 wild-type 1 cdc 1 marker 1 cdc & marker 1 wild-type pp. 46-51, 1974