* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Nature Rev.Mol.Cell Biol
Biology and consumer behaviour wikipedia , lookup
X-inactivation wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Point mutation wikipedia , lookup
Non-coding DNA wikipedia , lookup
Transgenerational epigenetic inheritance wikipedia , lookup
Molecular cloning wikipedia , lookup
DNA methylation wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Microevolution wikipedia , lookup
Designer baby wikipedia , lookup
History of genetic engineering wikipedia , lookup
Neocentromere wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Primary transcript wikipedia , lookup
Epigenetics wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epigenomics wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
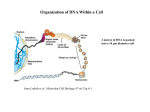
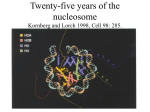
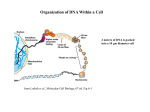
Organization of DNA Within a Cell 2 meters of DNA is packed into a 10 mm diameter cell from Lodish et al., Molecular Cell Biology, 6th ed. Fig 6-1 Appearance of Chromatin Depends on Salt Concentration Physiological ionic strength 30 nm fiber Low ionic strength Beads on a string from Lodish et al., Molecular Cell Biology, 6th ed. Fig 6-28 Nucleosomes are Packaged into a 30 nm Fiber from Lodish et al., Molecular Cell Biology, 6th ed. Fig 6-30 Chromatin Organization Each layer of chromatin organization reflects aspects of gene regulation Scaffold-associated regions can act as boundaries Condensed chromosomes are visible during metaphase from Zhou et al., Nature Rev.Mol.Cell Biol. 12, 7 (2011) Genes Can be Localized on Drosophila Polytene Chromosomes Polytene chromosomes exhibit a characteristic banding pattern Localization of a gene by in situ hybridization Biotinylated probe was detected by avidin conjugated to alkaline phosphatase AP substrate results in the formation of an insoluble precipitate at the site of hybridization from Lodish et al., Molecular Cell Biology, 6th ed. Fig 6-44 Chromosome Puffs Ecdysone produces a characteristic pattern of puffs in polytene chromosomes of salivary glands Puffs correspond to actively transcribed genes from Alberts et al., 3rd ed., Fig. 8-23 Actively Transcribed Genes are Present in Decondensed Chromatin Loss of 4.6 kb Bam HI fragment when the b-globin gene is active and histones are acetylated The 4.6 kb Bam HI fragment is present when the b-globin gene is inactive and histones are deacetylated from Lodish et al., Molecular Cell Biology, 6th ed. Fig 6-32 Nucleosome Structure Nucleosomes contain 2 copies of H2A, H2B, H3 and H4 147 bp of DNA is wrapped around nucleosome Histone tails emanate from core Some nucleosomes contain histone variants H1 is a linker histone from Jiang and Pugh, Nature Rev.Genet. 10, 161 (2009) Assembly of Nucleosomes Histone chaperones assemble histones into nucleosomes Histone chaperones prevent non-specific associations of histones with DNA Histone chaperones prevent formation of deleterious off-pathway intermediates from Das et al., Trends Biochem.Sci. 35, 476 (2010) Histone Tails Histones contain flexible termini that extend from the globular structure of the nucleosome from Lodish et al., Molecular Cell Biology, 6th ed. Fig 6-31 Modification of Histone Tails Chromatin structure is a source of epigenetic information Posttranslational modifications and histone variants contribute to structural and functional characteristics of chromatin The combination of histone modifications constitutes the histone code from Lodish et al., Molecular Cell Biology, 6th ed. Fig 6-31 The histone code influences chromatin condensation and function and defines actual or potential transcription states Histone Modifications Affect Chromatin Structure H3K4 methylation and H3K9 acetylation are hallmarks of active chromatin H3K27 methylation and H3K9 methylation are hallmarks of silent chromatin from Johnstone and Baylin, Nature Rev.Genet. 11, 806 (2010) Histone Modifications Define Functional Elements Each histone modification has a unique biological role Histone modifications are interdependent from Zhou et al., Nature Rev.Mol.Cell Biol. 12, 7 (2011) Chromatin Immunoprecipitation (ChIP) Use antibody to acetylated histone tail to determine the acetylation state of chromatin Antibody against any DNA binding protein determines the location of the binding site from Lodish et al., Molecular Cell Biology, 6th ed. Fig 7-37 Nucleosome Position Can Be Mapped Nucleosome position can be mapped by ChIP-seq Cross-link histones to DNA and digest linker Immunoprecipitate Sequence DNA from Jiang and Pugh, Nature Rev.Genet. 10, 161 (2009) Positioning of Nucleosomes at Promoters Nucleosome-free region at the beginning and end of genes Nucleosomes have defined locations near the promoter from Jiang and Pugh, Nature Rev.Genet. 10, 161 (2009) Nucleosome position is determined by ATP-dependent trans-acting factors Properties of Acetylated Histones Less positively charged Chromatin is less condensed H4K16Ac prevents formation of 30 nm fiber Control of Gene Expression by Acetylation Repressor recruits a complex that contains a histone deacetylase Neighboring histones are deacetylated Activator recruits a complex that contains a histone acetylase Neighboring histones are acetylated from Lodish et al., Molecular Cell Biology, 6th ed. Fig 7-38 Effect of Histone H3 K9 Methylation SUV39 methylates K9 Methylated K9 recruits HP1 Heterochromatin formation HP1 binds to SUV39 to propagate methylation Methylated K9 or phosphorylated S10 inhibits methylation of K9 from Turner, Cell 111, 285 (2002) Effect of Histone H3K4 Methylation Set9 methylates K4 Inhibits association of NuRD remodeling and deacetylase complex Inhibits association of SUV39 H3K4me is associated with active genes from Turner, Cell 111, 285 (2002) BRCA1 Modifies Pericentric Heterochromatin BRCA1 promotes enrichment of Ub-H2A in pericentric heterochromatin Loss of BRCA1 triggers transcription of satellite-DNA in pericentric heterochromatin Satellite-DNA transcription is sufficient to induce genome instability after loss of BRCA1 from Venkitaraman, Nature 477, 169 (2011) Nucleosomal Histones and Their Variants from Sarma and Reinberg, Nature Rev.Mol.Cell Biol. 6, 139 (2005) Role of H2A.Z H2A.Z is found on either side of a nucleosome-free region H2A.Z may protect promoters from being methylated from Talbert and Henikoff, Nature Rev.Mol.Cell Biol. 11, 264 (2010) Action of HMG-box Proteins HMG-box proteins bend DNA DNA bending can affect transcription and site-specific recombination from Thomas and Travers, Trends Biochem.Sci. 26, 167 (2001) Spatial Assembly of Expression Units from Dekker, Science 319, 1793 (2008) Regulation of DNA Accessibility Nucleosome sliding exposes binding sites Chromatin remodelling complexes extract DNA from the nucleosome surface Nucleosome eviction may be necessary for transcription initiation from Jiang and Pugh, Nature Rev.Genet. 10, 161 (2009) Histone chaperones incorporate histone variants