* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download GSEA - Bioinformatics Unit
Genomic imprinting wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Point mutation wikipedia , lookup
Metagenomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Copy-number variation wikipedia , lookup
Genetic engineering wikipedia , lookup
Pathogenomics wikipedia , lookup
Public health genomics wikipedia , lookup
Genome evolution wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Helitron (biology) wikipedia , lookup
The Selfish Gene wikipedia , lookup
Gene therapy wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene desert wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene nomenclature wikipedia , lookup
Microevolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
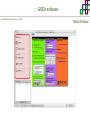
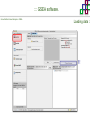
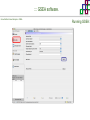
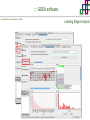
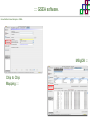
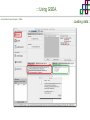
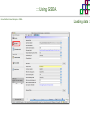
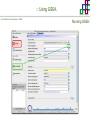
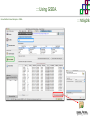
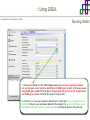
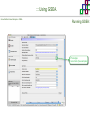
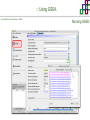
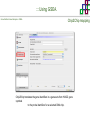
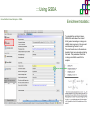
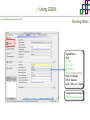
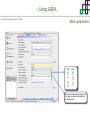
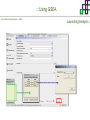
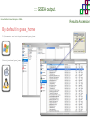
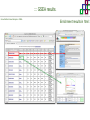
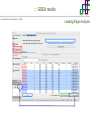
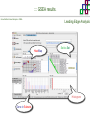
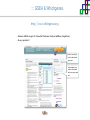
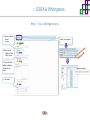
Course on Functional Analysis ::: Gene Set Enrichment Analysis - GSEA Madrid, Feb 16th, 2009. Gonzalo Gómez, PhD. [email protected] Bioinformatics Unit CNIO ::: Contents. 1. 2. 3. 4. 5. 6. 7. Introduction. GSEA Software Data Formats Using GSEA GSEA Output GSEA Results Leading Edge Analysis ::: Contents. 1. 2. 3. 4. 5. 6. 7. Introduction. GSEA Software Data Formats Using GSEA GSEA Output GSEA Results Leading Edge Analysis ::: Introduction. Gene Set Enrichment Analysis - GSEA - GSEA MIT Broad Institute v 2.0 available since Jan 2007 v 2.0.1 available since Feb 16th 2007 Version 2.0 includes Biocarta, Broad Institute, GeneMAPP, KEGG annotations and more... Platforms: Affymetrix, Agilent, CodeLink, custom... (Subramanian et al. PNAS. 2005.) ::: Introduction. Gene Set Enrichment Analysis - GSEA - ::: How works GSEA GSEA applies Kolmogorov-Smirnof test to find assymmetrical distributions for defined blocks of genes in datasets whole distribution. Is this particular Gene Set enriched in my experiment? Genes selected by researcher, Biocarta pathways, GeneMAPP sets, genes sharing cytoband, genes targeted by common miRNAs …up to you… ::: Introduction. Gene Set Enrichment Analysis - GSEA - ::: K-S test The Kolmogorov–Smirnov test is used to determine whether two underlying one-dimensional probability distributions differ, or whether an underlying probability distribution differs from a hypothesized distribution, in either case based on finite samples. The one-sample KS test compares the empirical distribution function with the cumulative distribution functionspecified by the null hypo The main applications are testing goodness of fit with the normal and uniform distributions. The two-sample KS test is one of the most useful and general nonparametric methods for comparing two samples, as it is sensitive to in both location and shape of the empirical cumulative distribution functions of the two samples. Dataset distribution Number of genes Gene set 1 distribution Gene set 2 distribution Gene Expression Level ::: Introduction. Gene Set Enrichment Analysis - GSEA - ClassA ::: How works GSEA ClassB FDR<0.05 ...testing genes independently... ttest cut-off FDR<0.05 Biological meaning? ::: Introduction. Gene Set Enrichment Analysis - GSEA - ::: How works GSEA - ClassA ClassB Gene Set 1 Gene Set 2 Gene Set 3 Gene set 3 enriched in Class B ES/NES statistic ttest cut-off Gene set 2 enriched in Class A + ::: Introduction. Gene Set Enrichment Analysis - GSEA - ES examples :: ::: Introduction. Gene Set Enrichment Analysis - GSEA - The Enrichment Score NES pval FDR Benjamini-Hochberg ::: Contents. 1. 2. 3. 4. 5. 6. 7. Introduction. GSEA Software Data Formats Using GSEA GSEA Output GSEA Results Leading Edge Analysis ::: GSEA software. Gene Set Enrichment Analysis - GSEA - Download :: http://www.broad.mit.edu/gsea/ ::: GSEA software. Gene Set Enrichment Analysis - GSEA - Main Window :: ::: GSEA software. Gene Set Enrichment Analysis - GSEA - Loading data :: !!! ::: GSEA software. Gene Set Enrichment Analysis - GSEA - Running GSEA : ::: GSEA software. Gene Set Enrichment Analysis - GSEA - Leading Edge Analysis ::: GSEA software. Gene Set Enrichment Analysis - GSEA - MSigDB ::: Chip to Chip Mapping ::: ::: Contents. 1. 2. 3. 4. 5. 6. 7. Introduction. GSEA Software Data Formats Using GSEA GSEA Output GSEA Results Leading Edge Analysis ::: Data Formats. Gene Set Enrichment Analysis - GSEA - ::: Data Formats. Gene Set Enrichment Analysis - GSEA - ::: Data Formats. Gene Set Enrichment Analysis - GSEA - Expression datasets *.gct ::: Data Formats. Gene Set Enrichment Analysis - GSEA - Expression datasets *.res ::: Data Formats. Gene Set Enrichment Analysis - GSEA - Expression datasets *.pcl ::: Data Formats. Gene Set Enrichment Analysis - GSEA - Expression datasets *.txt ::: Data Formats. Gene Set Enrichment Analysis - GSEA - Phenotype datasets *.cls For categorical phenotypes (e.g. Tumor vs Control) ::: Data Formats. Gene Set Enrichment Analysis - GSEA - Phenotype datasets For continuous phenotypes (e.g. Gene correlated to GeneSet) Time serie (each 30 minutes) Peak profile wanted For continuous phenotypes (e.g. Gene vs Time Series) ::: Data Formats. Gene Set Enrichment Analysis - GSEA - Gene Set Database *.gmx ::: Data Formats. Gene Set Enrichment Analysis - GSEA - Gene Set Database *.gmt ::: Data Formats. Gene Set Enrichment Analysis - GSEA - Ranked list format :: *.rnk ::: Contents. 1. 2. 3. 4. 5. 6. 7. Introduction. GSEA Software Data Formats Using GSEA GSEA Output GSEA Results Leading Edge Analysis ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Loading data :: ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Loading data :: ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Running GSEA : ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - ::: MSigDB. gsea_home ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Running GSEA : 1. Choose true (default) to have GSEA collapse each probe set in your expression dataset into a single gene vector, which is identified by its HUGO gene symbol. In this case, you are using HUGO gene symbols for the analysis. The gene sets that you use for the analysis must use HUGO gene symbols to identify the genes in the gene sets. 2. Choose false to use your expression dataset "as is." In this case, you are using the probe identifiers that are in your expression dataset for the analysis. The gene sets that you use for the analysis must also use these probe identifiers to identify the genes in the gene sets. ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Running GSEA : Phenotype Gene Sets (few samples) ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Running GSEA : ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Chip2Chip mapping Chip2Chip translates the gene identifiers in a gene sets from HUGO gene symbols to the probe identifiers for a selected DNA chip. ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Enrichment statistic : To calculate the enrichment score, GSEA first walks down the ranked list of genes increasing a running-sum statistic when a gene is in the gene set and decreasing it when it is not. The enrichment score is the maximum deviation from zero encountered during that walk. This parameter affects the running-sum statistic used for the analysis. ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Ranking Metric : Signal2Noise tTest Cosine Euclidean Manhatten Pearson (time series) Ratio of Classes Diff of Classes Log2_Ratio_of_Classes Categorical phenotypes Continuous phenotypes ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Ranking Metric : ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Ranking Metric : ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - More parameters : real abs 8.2 8.1 8.0 … -7.5 -7.7 -7.9 8.2 8.1 8.0 7.9 7.7 7.5 … parameter to determine whether to sort the genes in descending (default) or ascending order. ::: Using GSEA. Gene Set Enrichment Analysis - GSEA - Launching Analysis :: ::: Contents. 1. 2. 3. 4. 5. 6. 7. Introduction. GSEA Software Data Formats Using GSEA GSEA Output GSEA Results Leading Edge Analysis ::: GSEA output. Gene Set Enrichment Analysis - GSEA - By default in gsea_home C:\Documents and settings\username\gsea_home /Users/yourhome/gsea_home Results Accession ::: Contents. 1. 2. 3. 4. 5. 6. 7. Introduction. GSEA Software Data Formats Using GSEA GSEA Output GSEA Results Leading Edge Analysis ::: GSEA results. Gene Set Enrichment Analysis - GSEA - Index.html ::: Heat map of the top 50 features for each phenotype and a plot showing the correlation between the ranked genes and the phenotypes. In a heat map, expression values are represented as colors, where the range of colors (red, pink, light blue, dark blue) shows the range of expression values (high, moderate, low, lowest). ::: GSEA results. Gene Set Enrichment Analysis - GSEA - Enrichment results in html : ::: GSEA results. Gene Set Enrichment Analysis - GSEA - Enrichment results in html : ::: GSEA results. Gene Set Enrichment Analysis - GSEA - Enrichment results in html : How can I decide about my results? FDR ≤ 0.25 NOM p-val ≤ 0.05 ::: Contents. 1. 2. 3. 4. 5. 6. 7. Introduction. GSEA Software Data Formats Using GSEA GSEA Output GSEA Results Leading Edge Analysis ::: GSEA results. Gene Set Enrichment Analysis - GSEA - Leading Edge Analysis ::: GSEA results. Gene Set Enrichment Analysis - GSEA - Leading Edge Analysis HeatMap Set-to-Set Histogram Gene in Subsets ::: GSEA results. Gene Set Enrichment Analysis - GSEA - Leading Edge Analysis Heat Map The heat map shows the (clustered) genes in the leading edge subsets. In a heat map, expression values are represented as c where the range of colors (red, pink, light blue, dark blue) shows the range of expression values (high, moderate, low, lowest). ::: GSEA results. Gene Set Enrichment Analysis - GSEA - Leading Edge Analysis Set-to-Set The graph uses color intensity to show the overlap between subsets: the darker the color, the greater the overlap between the su When you compare a leading edge subset to itself, its members completely overlap so the corresponding cell is dark green. When you compare two subsets that have no overlapping members, the corresponding cell is white. ::: GSEA results. Gene Set Enrichment Analysis - GSEA - Leading Edge Analysis Gene in Subsets The graph shows each gene and the number of subsets in which it appears. ::: GSEA results. Gene Set Enrichment Analysis - GSEA - Leading Edge Analysis Histogram The last plot is a histogram, where the Jacquard is the intersection divided by the union for a pair of leading edge subsets. Number of Occurrences is the number of leading edge subset pairs in a particular bin. In this example, most subset pairs have no overlap (Jacquard = 0). ::: GSEA & FatiScan. Gene Set Enrichment Analysis - GSEA - Detects significant functions with Gene Ontology InterPro motifs, Swissprot KW and KEGG pathways in lists of genes ordered according to differents characteristics. ::: GSEA & Whichgenes. http://www.whichgenes.org - Retrieve miRNAs targets for Gene Set Enrichment Analysis (miRBase, TargetScan) - Always updated ! Enter if you simply want to download gene sets. Login whether you want to download and store your gene sets ::: GSEA & Whichgenes. http://www.whichgenes.org Create Sets 1. Choose oraanism. -Human - Mouse Looking for examples ? 2. Select source: - miRBase, TScan - Other sources 3. Copy and paste miRNAs identifiers. Create set per items. 4. Job name. Retrieving targets ::: GSEA & Whichgenes. http://www.whichgenes.org Gene Sets Cart 1. Choose gene sets for downloading. 2. Select output format. e.g. .CSV, .TSV, .gmt, .gmx 3. Select identifier. e.g. Agilent, Affy, Mgi… 4. DOWNLOAD GENE SETS !!! T H A N K [email protected] S