* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Identification and Molecular Characterization of Circadian Clock
Quantitative trait locus wikipedia , lookup
RNA silencing wikipedia , lookup
Pathogenomics wikipedia , lookup
Public health genomics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
RNA interference wikipedia , lookup
Gene nomenclature wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene desert wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Genetically modified crops wikipedia , lookup
Minimal genome wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genomic imprinting wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome (book) wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genome evolution wikipedia , lookup
Gene expression programming wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Designer baby wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
History of genetic engineering wikipedia , lookup
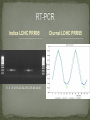
By Matthew Geniza Mentors: Dr. Todd Mockler and Dr. Sergei Filichkin To provide a foundation of the molecular mechanisms that determine plant adaptations depending on their environment. To increase the basic knowledge of how transcription factors relate to phase-specific gene expression depending on the external conditions. If Myb-10 is a clock gene, over expressing or suppressing it alters the expression of other clock or clock-regulated genes. The plant circadian clock - at least three interlinked loops PRR5 PRR7 PRR9 INPUT CCA1 LHY light OUTPUT flowering germination growth temperature X Y GI photosynthesis metabolism TOC1 LUX Plants integrate daily temperature and light information into the circadian clock in order to gauge the time of day and season, and thus coordinate biological processes appropriately for the environment. Plants have evolved an endogenous circadian clock Internal biological processes are synchronized with changes in the environment. RNA transcripts of diurnal and circadian regulated genes peak at certain times in the day and night. Understanding the regulation of gene expression is important in the event of major climate changes Rice is crop plant that serves as a stable food for a majority of the world. Rice has a sequenced and annotated genome Is Myb-10 a clock gene? Does it control oscillations of mRNAs of other rice genes? Does it affect the phases of other clock genes? Will phase shifting caused by Myb-10 mis-expression changes in phenotype, i.e. growth rate, flowering timing, and ultimately – seed production? If yes, then Myb-10 is a clock-associated regulator. Normalized Expression 1.2 A. thaliana Rice Myb10 GeneID: Os05g34110 Poplar Putative Myb-like TF AT5G05090 1 0.8 0.6 0.4 0.2 0 0 4 8 12 16 20 24 28 32 36 40 Phase http://diurnal.cgrb.oregonstate.edu/ 44 If the Myb-10 gene is a core clock gene, then it will change the oscillations of other core clock genes. If the Myb-10 gene is NOT a core clock gene, then it will change oscillations of other diurnally regulated genes, but not core clock genes. To identify diurnally regulated genes in rice that are affected by over expression or silencing of a specific circadian clock regulated transcription factor. To identify cis elements in the promoter region of Myb-10 which regulate its diurnal oscillations. Rice plants are grown in 12 hours of light and 12 hours of dark cycle for 7 days. This is followed by 60 hours of continuous light. Tissue samples from plants are collected every 4 hours during the continuous light period. RNA isolation Bioanalyzer Assayed by RT PCR to determine cycling profiles of diurnally regulated gene targets Microarray The cycling of transcripts of several clock genes (such as CIRCADIAN CLOCK ASSOCIATED 1) is studied in the wild type rice and transgenic plants expressing or suppressing Myb-10. RIN 10.0 RIN 7.0 Gel Image Indica LDHC CCA1 0 4 8 12 16 20 24 28 32 36 40 44 48 Diurnal LDHC CCA1 Indica LDHC PRR95 0 4 8 12 16 20 24 28 32 36 40 44 48 Diurnal LDHC PRR95 Quantitative data Measures gene expression Assays all genes Analyze Microarray Data for Indica Prepare transgenic Myb-10 plants for Microarray Analyze Microarray Data for Myb-10 plants Compare Indica and transgenic Myb-10 plants Dr. Todd Mockler Dr. Sergei Filichkin The Mockler Lab Dr. Kevin Ahern HHMI