* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Other RNA Processing Events
Artificial gene synthesis wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome evolution wikipedia , lookup
Microevolution wikipedia , lookup
Metagenomics wikipedia , lookup
Genetic code wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Expanded genetic code wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
RNA interference wikipedia , lookup
RNA silencing wikipedia , lookup
Polyadenylation wikipedia , lookup
RNA-binding protein wikipedia , lookup
History of RNA biology wikipedia , lookup
Messenger RNA wikipedia , lookup
Transfer RNA wikipedia , lookup
Primary transcript wikipedia , lookup
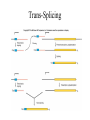
Other RNA Processing Events Chapter 16 Ribosomal RNA Processing • rRNA genes of both eukaryotes and bacteria are transcribed as larger precursors must be processed to yield rRNAs of mature size - Several different rRNA molecules are embedded in a long precursor and each must be cut out Eukaryotic rRNA processing • The rRNA are separated by regions called nontranscribed spacers (NTSs). • Transcribed spacers - regions of the gene that are transcribed as part of rRNA precursor - and then removed in processing of precursor to mature rRNA species. Eukaryotic rRNA Processing • Ribosomal RNAs are made in eukaryotic nucleoli as precursors that must be processed to release mature rRNAs • Order of RNAs in the precursor is (fig: 16.2) – – – – 18S 5.8S 28S in all eukaryotes Exact sizes of the mature rRNAs vary from one species to another Eukaryotic rRNA Processing • rRNA processing – nucleolus – small nucleolar RNAs (snoRNAs) + proteins = Small nucleolar ribonucleoproteins (snoRNPs) • E1, E2 and E3 – interact with distinct regions in prerRNA Bacterial rRNA Processing • Bacterial rRNA precursors contain tRNA and all 3 rRNA • rRNA are released from their precursors by RNase III and RNase E – RNase III is the enzyme that performs at least the initial cleavages that separate the individual large rRNAs – RNase E is another ribonuclease that is responsible for removing the 5S rRNA from the precursor Processing Bacterial rRNA Transfer RNA Processing • Transfer RNAs are made in all cells as overly long precursors – These must be processed by removing RNA at both ends • Nuclei of eukaryotes contain precursors of a single tRNA • In bacteria - precursor may contain one or more tRNA - Sometimes a mixture of rRNAs and tRNAs Cutting apart Polycistronic Precursors • In processing bacterial RNA that contain more than one tRNA – First step is to cut precursor up into fragments with just one tRNA each • Cutting between tRNAs in precursors having 2 or more tRNA • Cutting between tRNAs and rRNAs in precursors – Enzyme that performs both chores is the RNase III Forming Mature 5’-Ends of tRNA • Extra nucleotides are removed from the 5’ends of pre-tRNA in one step by an endonucleolytic cleavage catalyzed by RNase P • RNase P from bacteria and eukaryotic nuclei have a catalytic RNA subunit called M1 RNA Processing 3’-Ends of tRNA RNase D, RNase BN, RNase T, RNase PH, RNase II and polynucleotide phosphorylase (PNPase) Forming mature 3’-Ends of tRNA • RNase II and polynucleotide phosphorylase cooperate – To remove most of extra nucleotides at the end of a tRNA precursor • RNases PH and T are most active in removing the last 2 nucleotides from RNA – RNase T is the major participant in removing very last nucleotide Trans-Splicing • Splicing that occurs in all eukaryotic species is called cis-splicing because it involves 2 or more exons that exist together in the same gene • trans-splicing has exons that are not part of the same gene at all - may not even be on the same chromosome The Mechanism of Trans-Splicing • Trans-splicing occurs in several organisms – Parasitic and free-living worms – First discovered in trypanosomes • Trypanosome mRNA are formed by transsplicing between a short leader exon and any one of many independent coding exons Trans-Splicing Trans-Splicing Scheme • Branchpoint adenosine within the half-intron attached to the coding exon attacks the junction between the leader exon and its half-intron • Creates a Y-shaped intron-exon intermediate analogous to the lariat intermediate Mechanism of Editing • Unedited transcripts can be found along with edited versions of the same mRNAs • Editing occurs in the poly(A) tails of mRNAs that are added posttranscriptionally • Partially edited transcripts have been isolated always edited at their 3’-ends but not at their 5’-ends Role of gRNA in Editing • Guide RNAs (gRNA) could direct the insertion and deletion of UMPs over a stretch of nucleotides in the mRNA • When editing is done, gRNA could hybridize near the 5’-end of newly edited region Guide RNA Editing • 5’-end of the first gRNA hybridizes to an unedited region at the 3’-border of editing I the pre-mRNA • The 5’-ends of the rest of the gRNAs hybridize to edited regions progressively closer to the 5’end of the region to be edited in the premRNA • All of these gRNAs provide A’s and G’s as templates for the incorporation of U’s missing from the mRNA Mechanism of Removing U’s • Sometimes the gRNA is missing an A or G to pair with a U in the mRNA – In this case the U is removed • Mechanism of removing U’s involves – Cutting pre-mRNA just beyond U to be removed – Removal of U by exonuclease – Ligating the two pieces of pre-mRNA together • Mechanism of adding U’s uses same first and last step • Middle step involves addition of one or more U’s from UTP by TUTase RNA Interference • RNA interference occurs when a cell encounters dsRNA from: – Virus – Transposon – Transgene • Trigger dsRNA is degraded into 21-23 nt fragments (siRNAs) by an RNase III-like enzyme called Dicer • Double-stranded siRNA, with Dicer and Dicerassociated protein R2D2 form a complex called complex B Complex B • Complex B delivers the siRNA to the RISC loading complex (RLC) – Separates 2 strands of siRNA – Transfers guide strand to RNA-induced silencing complex (RISC) that introduces a protein- Ago2 Complex B • The guide strand of siRNA base-pairs with target mRNA in the active site of PIWI domain of Ago2 – Ago2 is an RNase H-like enzyme known as a slicer – Slicer cleaves the target mRNA in middle of the region of its base-pairing with the siRNA – ATP-dependent step has cleaved RNA ejected from RISC which then accepts a new molecule of mRNA for degradation • • • This project is funded by a grant awarded under the President’s Community Based Job Training Grant as implemented by the U.S. Department of Labor’s Employment and Training Administration (CB-15-162-06-60). NCC is an equal opportunity employer and does not discriminate on the following basis: against any individual in the United States, on the basis of race, color, religion, sex, national origin, age disability, political affiliation or belief; and against any beneficiary of programs financially assisted under Title I of the Workforce Investment Act of 1998 (WIA), on the basis of the beneficiary’s citizenship/status as a lawfully admitted immigrant authorized to work in the United States, or his or her participation in any WIA Title I-financially assisted program or activity. Disclaimer • This workforce solution was funded by a grant awarded under the President’s Community-Based Job Training Grants as implemented by the U.S. Department of Labor’s Employment and Training Administration. The solution was created by the grantee and does not necessarily reflect the official position of the U.S. Department of Labor. The Department of Labor makes no guarantees, warranties, or assurances of any kind, express or implied, with respect to such information, including any information on linked sites and including, but not limited to, accuracy of the information or its completeness, timeliness, usefulness, adequacy, continued availability, or ownership. This solution is copyrighted by the institution that created it. Internal use by an organization and/or personal use by an individual for non-commercial purposes is permissible. All other uses require the prior authorization of the copyright owner.