* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Inquiry into Life Twelfth Edition
Secreted frizzled-related protein 1 wikipedia , lookup
Genome evolution wikipedia , lookup
Genomic imprinting wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Gene expression profiling wikipedia , lookup
Molecular cloning wikipedia , lookup
Ridge (biology) wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Molecular evolution wikipedia , lookup
Gene expression wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gene regulatory network wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Acetylation wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Non-coding DNA wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Transcription factor wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Silencer (genetics) wikipedia , lookup
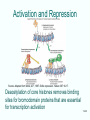
Lecture PowerPoint to accompany Molecular Biology Fourth Edition Robert F. Weaver Chapter 13 Chromatin Structure and Its Effects on Transcription Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. 13.1 Histones • Eukaryotic cells contain 5 kinds of histones – – – – – H1 H2A H2B H3 H4 • Each histone type isn’t homogenous – Gene reiteration – Posttranslational modification Source: Panyim and Chalkley, Arch. Biochem. 13-2 & Biophys. 130, 1969, f. 6A, p.343. Properties of Histones • Abundant proteins whose mass in nuclei nearly equals that of DNA • Pronounced positive charge at neutral pH • Most are well-conserved from one species to another • Not single copy genes, repeated many times – Some copies are identical – Others are quite different – H4 has only had 2 variants ever reported 13-3 13.2 Nucleosomes • Chromosomes are long, thin molecules that will tangle if not carefully folded • Folding occurs in several ways • First order of folding is the nucleosome – X-ray diffraction has shown strong repeats of structure at 100Å intervals – This spacing approximates the nucleosome spaced at 110Å intervals 13-4 Histones in the Nucleosome • Chemical cross-linking in solution: – H3 to H4 – H2A to H2B • H3 and H4 exist as a tetramer (H3-H4)2 • Chromatin is composed of roughly equal masses of DNA and histones – Corresponds to 1 histone octamer per 200 bp of DNA – Octamer composed of: • 2 each H2A, H2B, H3, H4 • 1 each H1 13-5 H1 and Chromatin • Treatment of chromatin with trypsin or high salt buffer removes histone H1 • This treatment leaves chromatin looking like “beads-on-a-string” • The beads named nucleosomes – Core histones form a ball with DNA wrapped around the outside – DNA on outside minimizes amount of DNA bending – H1 also lies on the outside of the nucleosome 13-6 Nucleosome Structure • Central (H3-H4)2 core attached to H2AH2B dimers • Grooves on surface define a left-hand helical ramp – a path for DNA winding – DNA winds almost twice around the histone core condensing DNA length by 6- to 7-X – Core histones contain a histone fold: • 3 a-helices linked by 2 loops • Extended tail of abut 28% of core histone mass • Tails are unstructured 13-7 The 30-nm Fiber • Second order of chromatin folding produces a fiber 30 nm in diameter – The string of nucleosomes condenses to form the 30-nm fiber in a solution of increasing ionic strength – This condensation results in another six- to seven-fold condensation of the nucleosome itself • Four nucleosomes condensing into the 30nm fiber form a zig-zag structure 13-8 Formation of the 30-nm Fiber • Two stacks of nucleosomes form a lefthanded helix – Two helices of polynucleosomes – Zig-zags of linker DNA • Role of histone H1? – 30-nm fiber can’t form without H1 – H1 crosslinks to other H1 more often than to core histones 13-9 Higher Order Chromatin Folding • 30-nm fibers account for most of chromatin in a typical interphase nucleus • Further folding is required in structures such as the mitotic chromosomes • Model favored for such higher order folding is a series of radial loops Source: Adapted from Marsden, M.P.F. and U.K. Laemmli, Metaphase chromosome structure: Evidence of a radial loop model. Cell 17:856, 13-10 1979. Relaxing Supercoiling in Chromatin Loops • When histones are removed, 30-nm fibers and nucleosomes disappear • Leaves supercoiled DNA duplex • Helical turns are superhelices, not ordinary double helix • DNA is nicked to relax 13-11 13.3 Chromatin Structure and Gene Activity • Histones, especially H1, have a repressive effect on gene activity in vitro • Two families of 5S rRNA genes studied are oocyte and somatic genes – Oocyte genes are expressed only in oocytes – Somatic genes are expressed both in oocytes and somatic cells – Somatic genes form more stable complexes with transcription factors 13-12 Transcription Factors and Histones Control the 5S rRNA • Genes active by TFIIIs preventing formation of nucleosome stable complexes with internal control region • Stable complexes require histone H1 and exclude TFIIIs once formed so that genes are repressed 13-13 Effects of Histones on Transcription of Class II Genes • Core histones assemble nucleosome cores on naked DNA • Transcription of reconstituted chromatin with an average of 1 nucleosome / 200 bp DNA exhibits 75% repression relative to naked DNA • Remaining 25% is due to promoter sites not covered by nucleosome cores 13-14 Histone H1 and Transcription • Histone H1 causes further repression of template activity, in addition to that of core histones • H1 repression can be counteracted by transcription factors • Sp1 and GAL4 act as both: – Antirepressors preventing histone repressions – Transcription activators • GAGA factor: – Binds to GA-rich sequences in the Krüppel promoter – An antirepressor – preventing repression by histones 13-15 Model of Transcriptional Activation Source: Adapted from Laybourn, P.J. and J. T. Kadonaga, Role of nucleosomal cores and histone H1 in regulation of transcription by polymerase II. Science 254:243, 1991. 13-16 Nucleosome Positioning • Model of activation and antirepression asserts that transcription factors can cause antirepression by: – Removing nucleosomes that obscure the promoter – Preventing initial nucleosome binding to the promoter • Both actions are forms of nucleosome positioning – activators force nucleosomes to take up positions around, not within, promoters 13-17 Nucleosome-Free Zones • Nucleosome positioning would result in nucleosome-free zones in the control regions of active genes • Assessment in a circular chromosome can be difficult without some type of marker 13-18 Detecting DNaseHypersensitive Regions • Active genes tend to have DNase-hypersensitive control regions • Part of this hypersensitivity is due to absence of 13-19 nucleosomes Histone Acetylation • Histone acetylation occurs in both cytoplasm and nucleus • Cytoplasmic acetylation carried out by HAT B (histone acetyltransferase, HAT) – Prepares histones for incorporation into nucleosomes – Acetyl groups later removed in nucleus • Nuclear acetylation of core histone N-terminal tails – Catalyzed by HAT A – Correlates with transcription activation – Coactivators of HAT A found which may allow loosening of association between nucleosomes and gene’s control region – Attracts bromodomain proteins, essential for transcription 13-20 Histone Deacetylation • Transcription repressors bind to DNA sites and interact with corepressors which in turn bind to histone deacetylases – Repressors • Unliganded nuclear receptors • Mad-Max – Corepressors • NCoR/SMRT • SIN3 – Histone deacetylases - HDAC1 and 2 13-21 Ternary Protein Complexes • Assembly of complex brings histone deacetylases close to nucleosomes • Deacetylation of core histones allows – Histone basic tails to bind strongly to DNA, histones in neighboring nucleosomes – This inhibits transcription 13-22 Activation and Repression Source: Adapted from Wolfe, A.P., 1997. Sinful repression. Nature 387:16-17. Deacetylation of core histones removes binding sites for bromodomain proteins that are essential for transcription activation 13-23 Chromatin Remodeling • Activation of many eukaryotic genes requires chromatin remodeling • Several protein complexes carry this out – All have ATPase harvesting energy from ATP hydrolysis for use in remodeling – Remodeling complexes are distinguished by ATPase component 13-24 Remodeling Complexes • SWI/SNF – In mammals, has BRG1 as ATPase – 9-12 BRG1-associated factors (BAFs) • A highly conserved BAF is called BAF 155 or 170 • Has a SANT domain responsible for histone binding • This helps SWI/SNF bind nucleosomes • ISWI – Have a SANT domain – Also have SLIDE domain involved in DNA binding 13-25 SWI/SNF Chromatin Remodeling 13-26 Mechanism of Chromatin Remodeling • Mechanism of chromatin remodeling involves: – Mobilization of nucleosomes – Loosening of association between DNA and core histones • Catalyzed remodeling of nucleosomes involves formation of distinct conformations of nucleosomal DNA/core histones when contrasted with: – Uncatalyzed DNA exposure in nucleosomes – Simple nucleosome sliding along a DNA stretch 13-27 Remodeling in Yeast HO Gene Activation • Chromatin immunoprecipitation (ChIP) can reveal the order of binding of factors to a gene during activation • As HO gene is activated: – First factor to bind is Swi5 – Followed by SWI/SNF and SAGA containing HAT Gcn5p – Next general transcription factors and other proteins bind • Chromatin remodeling is among the first steps in activation of this gene • Order could be different in other genes 13-28 Chromatin Immunoprecipitation 13-29 Remodeling in the Human IFN-b Gene: The Histone Code The Histone Code: – The combination of histone modifications on a given nucleosome near a gene’s control region affects efficiency of that gene’s transcription – This code is epigenetic, not affecting the base sequence of DNA itself • Activators in the IFN-b enhanceosome can recruit a HAT (GCN5) – HAT acetylates some Lys on H3 and H4 in a nucleosome at the promoter – Protein kinase phosphorylates Ser on H3 – This permits acetylation of another Lys on H3 13-30 Remodeling in the Human IFN-b Gene: TF Binding • Remodeling allows TFIID to bind 2 acetylated Lys in the nucleosomes through the dual bromodomain in TAFII250 • TFIID binding – Bends the DNA – Moves remodeled nucleosome aside – Paves the way for transcription to begin 13-31 Heterochromatin • Euchromatin: relatively extended and open chromatin that is potentially active • Heterochromatin: very condensed with its DNA inaccessible – Microscopically appears as clumps in higher eukaryotes – Repressive character able to silence genes as much as 3 kb away 13-32 Heterochromatin and Silencing • Formation of at tips of yeast chromosomes (telomeres) with silencing of the genes is the telomere position effect (TPE) • Depends on binding of proteins – RAP1 to telomeric DNA – Recruitment of proteins in this order: • SIR3 • SIR4 • SIR2 13-33 SIR Proteins • Heterochromatin at other locations in chromosome also depends on the SIR proteins • SIR3 and SIR4 interact directly with histones H3 and H4 in nucleosomes – Acetylation of Lys 16 on H4 in nucleosomes prevents interaction with SIR3 – Blocks heterochromatin formation • Histone acetylation also works in this way to promote gene activity 13-34 Histone Methylation • Methylation of Lys 9 in N-terminal tail of H3 attracts HP1 • This recruits a histone methyltransferase – Methylates Lys 9 on a neighboring nucleosome – Propagates the repressed, heterochromatic state • Methylation of Lys and Arg side chains in core histones can have either repressive or activating effects 13-35 Modification Interactions • The modifications shown above the tail are activating – Ser phosphorylation – Lys acetylation • Modification below the tail (Lys methylations) is repressive 13-36 Modification Combinations • Methylations occur in a given nucleosome in combination with other histone modifications: – Acetylations – Phosphorylations – Ubiquitylations • Each particular combination can send a different message to the cell about activation or repression of transcription • One histone modification can also influence other, nearby modifications 13-37 Nucleosomes and Transcription Elongation • An important transcription elongation facilitator is FACT (facilitates chromatin transcription) – Composed of 2 subunits: • Spt16 – Binds to H2A-H2B dimers – Has acid-rich C-terminus essential for these nucleosome remodeling activities • SSRP1 binds to H3-H4 tetramers – Facilitates transcription through a nucleosome by promoting loss of at least one H2A-H2B dimer from the nucleosome • Also acts as a histone chaperone promoting readdition of H2A-H2B dimer to a nucleosome that 13-38 has lost such a dimer