* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Transcription - smithlhhsb121
Transcription factor wikipedia , lookup
DNA supercoil wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
RNA interference wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Peptide synthesis wikipedia , lookup
Non-coding DNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
RNA silencing wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Metalloprotein wikipedia , lookup
Proteolysis wikipedia , lookup
Point mutation wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Silencer (genetics) wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Polyadenylation wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Biochemistry wikipedia , lookup
Gene expression wikipedia , lookup
Messenger RNA wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Genetic code wikipedia , lookup
Transfer RNA wikipedia , lookup
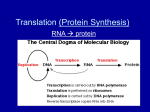
The replication of DNA takes place in S phase of interphase However, DNA is also used during G1 to assemble proteins This process is broken down into two distinct segments: transcription and translation The entire human genome is found in every cell, but only a portion is activated This portion has to convey the message outside the nucleus It is not possible to translate a gene directly to a protein RNA has to be used as an intermediary Quite similar to DNA, with a few key differences Structurally, it contains a ribose sugar ◦ This has a hydroxyl group in the 2’ position instead of a hydrogen Additionally, RNA tends to be single stranded, and contains uracil in the place of thymine DNA is not capable of leaving the nucleus A complement of one of the strands is transcribed as RNA ◦ Called messenger RNA (mRNA) RNA polymerase (II in eukaryotes) serves the duel purpose of splitting the strand and attaching the complementary nucleotides ◦ Again, in the 5’->3’ direction This binding is not random A region of DNA, called the promoter, initiates transcription The order of nucleotides in the promoter determines which strand gets transcribed A series of proteins, called transcription factors, bind to the promoter ◦ For example, a TATA box is a part of the promoter region The group of TFs and RNA pol is called a transcription initiation complex Transcription proceeds until a terminator is reached The actual detachment process is still a bit “iffy” The form of mRNA detaching from the DNA is not ready to be read yet It must go through two processes 1. Alteration of endings 2. Splicing of regions The 5’ end gets “capped” by a modified version of guanine This 5’ cap holds the pre-mRNA together and acts as a recognition molecule for ribosomes On the 3’ end, a region of 30 to 200 adenine nucleotides is added – called a poly(A) tail ◦ Again, to assist in holding the pre-mRNA together A large portion of the pre-mRNA will not leave the nucleus Portions are cut out, and the flanking regions are joined together The regions that are kept are exons, ones that are not are introns Short regions of nucleotides at the end of introns signal their removal Small nuclear ribonucleoproteins, snRNPs, join up with other proteins to form a spliceosome ◦ This carries out the splicing process ◦ http://www.youtube.com/watch?v=WsofH466lqk&f eature=related Now the mRNA is ready to be translated Recall that the message is “read” three nucleotides at a time This is the codon triplet The codon is interpreted by transfer RNA (tRNA) Each version has a nucleotide triplet of its own This serves as the complement to the codon of mRNA ◦ Hence, it is the anticodon All 64 permutations are covered by tRNA molecules, carrying the 20 different amino acids ◦ However, there are only about 45 difference tRNA molecules In many cases, the first two nucleotides are sufficient for recognition However, each tRNA carries one, and only one, amino acid The attachment of tRNA to amino acid is mediated by an aminoacyl-tRNA synthetase enzyme ◦ 20 different versions, one for each amino acid The interaction of tRNA and mRNA takes place in a ribosome Consists of two protein subunits and ribosomal RNA (rRNA) Within the ribosome are three binding sites ◦ P site (peptidyl-tRNA site) where the tRNA holding the polypepetide chain ◦ A site (aminoacyl-tRNA site) where next tRNA in line is held ◦ E site (exit site) where used tRNAs are discharged Begins with initiation Small subunit of ribosome attaches to leader segment of mRNA Initiator tRNA bearing methionine then attaches, followed by attachment of large ribosomal subunit Although methionine is initially attached, it is sometimes removed The next stage, elongation, is now set This is broken into three distinct steps 1. Codon recognition – mRNA codon hydrogen bonds with tRNA anticodon in A site 2. Peptide bond formation – rRNA molecule catalyzes formation of peptide bond (amino acid-amino acid bond) between adjacent amino acids in A and P sites 3. Translocation – amino acid in A site is moved to P site, and is now carrier of growing polypeptide chain (AA in P site moved to E site for removal) http://www.youtube.com/watch?v=5bLEDdPSTQ&feature=related Translation is finalized during termination Three stop codon triplets UAA, UAG and UGA, do not code for any amino acid Rather, a protein called a release factor binds to stop codon, causes a water molecule to attach The polypeptide chain is released and is ready for the folding process that will make it a functional protein http://www.youtube.com/watch?v=SMtWvDbfHLo&feature=relmfu http://www.youtube.com/watch?v=TfYf_rPWUdY&feature=relmfu