* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download DNA, RNA and Protein
Ancestral sequence reconstruction wikipedia , lookup
Deoxyribozyme wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Molecular evolution wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Citric acid cycle wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Protein adsorption wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Peptide synthesis wikipedia , lookup
Metalloprotein wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Point mutation wikipedia , lookup
Polyadenylation wikipedia , lookup
Protein structure prediction wikipedia , lookup
Proteolysis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Gene expression wikipedia , lookup
Non-coding RNA wikipedia , lookup
Biochemistry wikipedia , lookup
Bottromycin wikipedia , lookup
Messenger RNA wikipedia , lookup
Genetic code wikipedia , lookup
Expanded genetic code wikipedia , lookup
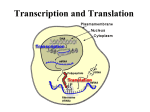
Translation How the Genetic Information Is Used to Build a Protein Information Flow From DNA replication DNA transcription initial transcript RNA processing mature RNA translation Protein Properties of the Genetic Code • Triplet – 3 nucleotides code for 1 amino acid • Non-overlapping – Codons are adjacent to each other • Degenerate – Some amino acids have more than one codon • “Almost” universal – A few exceptions occur in mitochondrial, bacterial and protist genes 5’3’ Sequence of CODONS Accurate Protein Synthesis Depends on Codon-Anticodon Recognition tRNA Charging • A specific tRNA synthetase catalyzes the attachment of the appropriate amino acid to each tRNA • Energy from conversion of ATP to AMP drives the reaction Overview of Translation At the ribosome, codons in mRNA are recognized by tRNA anticodons to place amino acids in the specific sequence determined by the DNA. Three Stages of Translation: Initiation- assemble components to start process Elongation- add amino acids in repeated cycles Termination- release protein product Initiation (Prokaryotic) Formation of initiation complex containing: Small ribosomal subunit mRNA Initiator (f-Met) tRNA Large ribosomal subunit f-met UAC 5’-------------AUGUUUCUCUGA---3’ mRNA Elongation a. next tRNA binds to mRNA at the A site E site P site A site f-met phe UAC AAA 5’ -------------AUGUUUCUCUGA---3’ mRNA Elongation b. amino acids are joined with peptidyl transferase peptide bond f-met phe UAC AAA 5’-------------AUGUUUCUCUGA---3’ mRNA Elongation c. ribosome moves by one codon (translocation) --growing peptide is now in P site --first tRNA is in E site f-met phe UAC AAA 5’-------------AUGUUUCUCUGA---3’ mRNA Elongation d. first tRNA is released from E site f-met UAC phe AAA 5’-------------AUGUUUCUCUGA---3’ mRNA Elongation (second cycle) a. next tRNA binds to mRNA at the A site f-met UAC phe leu AAA GAG 5’ -------------AUGUUUCUCUGA---3’ mRNA Elongation (second cycle) b. amino acids are joined with peptidyl transferase peptide bond UAC f-met phe leu AAA GAG 5’ -------------AUGUUUCUCUGA---3’ mRNA Elongation (second cycle) c. ribosome moves by one codon --growing peptide is now in P site --second tRNA is now in E site UAC f-met phe leu AAA GAG 5’-------------AUGUUUCUCUGA---3’ mRNA Elongation (second cycle) d. second tRNA is released from E site UAC f-met phe leu AAA GAG 5’-------------AUGUUUCUCUGA---3’ mRNA Termination a. release factor binds to stop codon UAC f-met AAA phe leu RF GAG 5’-------------AUGUUUCUCUGA---3’ mRNA Termination b. protein chain is released other components separate UAC f-met phe leu GAG Large Subunit RF Small Subunit AAA 5’-------------AUGUUUCUCUGA---3’ mRNA Applying Your Knowledge If the mRNA sequence for codons 5, 6, and 7 of a protein is 5’-AAG-AUU-GGA-3’, what is the amino acid sequence in the protein? 1. Gly-ile-lys 2. Arg-leu-glu 3. Glu-leu-arg 4. Asn-met-gly 5. Lys-ile-gly Control of Initiation in Prokaryotes A. Small Ribosomal Subunit binds to mRNA a. IF-3 binds to small ribosomal subunit b. Small ribosomal subunit binds to Shine-Dalgarno sequence on mRNA A sequence in the 16S rRNA of the small ribosomal subunit is complementary to the Shine-Dalgarno sequence on mRNA IF-3 = Initiation Factor 3 Control of Initiation in Prokaryotes B. Formyl-methionine tRNA binds to mRNA a. IF-2 +GTP + f-met-tRNA join b. f-met-tRNA binds to the first codon c. IF-1 joins to small subunit d. IFs dissociate, GTP is hydrolyzed to GDP C. Large Ribosomal Subunit binds to mRNA Control of Initiation in Eukaryotes Binding of Small Subunit assisted by proteins bound to -5’-methyl guanine cap -Poly-A tail Control of Elongation by Elongation Factors EF-Tu joins GTP and a charged tRNA to form a complex that binds to the A site EF-Ts regenerates the EF-Tu + GTP complex EF-G and GTP are required for ribosome translocation Control of Termination by Release Factors RF1 binds to UAA and UAG RF2 binds to UAA and UGA RF3 forms a complex with GTP that binds to the ribosome Energetics of Translation Number of Phosphate Bonds Required (use of ATP or GTP) Initiation 1 for assembly of ribosomal subunits 2 for activation of f-met tRNA Elongation 2 for activation of each tRNA 2 for addition of each amino acid Termination 1 for dissociation of ribosomal subunits Energetics of Translation How many phosphate bonds are required to build a protein of 100 amino acids? First Amino Acid Next 99 Amino Acids Termination Total 3 99x4=396 1 400